Rougeulle group
Non coding RNA, differentiation and development

© Rougeulle Team
Research in the Rougeulle lab focuses on the emblematic process of X chromosome inactivation, with the aim to elucidate how such a chromosome-wide epigenetic silencing is achieved and regulated in various mammalian species. More generally, we are interested in the epigenetic control of gene expression in relation to cell identity and fate, the contribution of long noncoding RNA genes and transposable elements to these processes and the plasticity of epigenetic regulations in evolution.
We use a variety of mouse and primate pluripotent stem cells and their differentiated derivatives to recapitulate key developmental stages and cellular states. Our experimental strategy combines genome engineering (CRISPR) with single cell analyses, large scale transcriptomic and epigenomic investigations and 3D topological studies.
Our research projects :
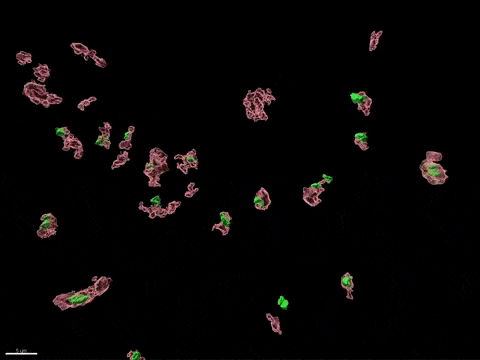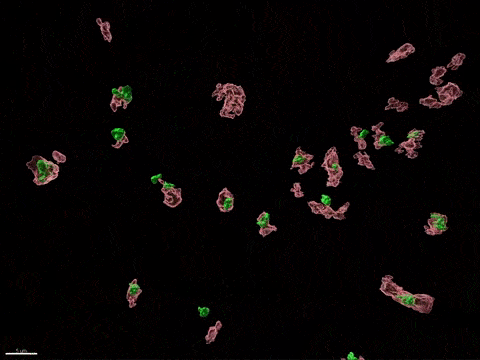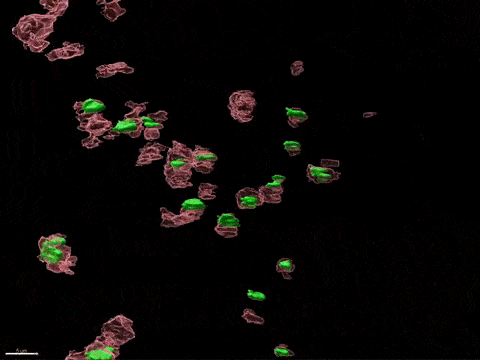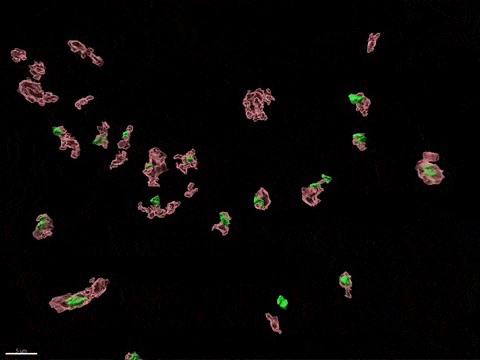
Human X-inactivation in a dish
Description
X chromosome inactivation is critical for women development and linked to sex-specific susceptibilities to various diseases. To understand how XCI contributes to male-female differences, we model, in vitro, key steps of X-inactivation using, as an entry point, Human Embryonic Stem cells (hESC). hESC are reprogrammed to discover how XCI is initially established during the first days of embryonic development with a particular focus on the role of structural noncoding RNAs such as XIST and XACT. hESC are differentiated into specific cell types to assess the plasticity of XCI in distinct somatic tissues, including the hematopoietic compartment and organized tissue through the use of organoids.

Evolution of X-inactivation across primates
Description
Remarkable differences exist in the way X chromosome inactivation is set up between human and mouse in connection with drastically different network of regulatory lncRNA genes which our group contributed to decipher. We are now exploring phenotypic and mechanistic variability in X-inactivation across primates using non-human primate pluripotent stem cells to probe the degree of genetic and epigenetic innovation across a limited timescale, and the contribution of the noncoding genome to such evolution.
Plasticity of the noncoding genome
Description
The noncoding genome participates to the establishment of gene expression programs, and, as such, to cell identity and fate. It encompasses a vast repertoire of lncRNA genes (LRGs) – a fraction of which derives from endogenous retroviruses – whose function may be conveyed by distinct modules, such as the RNA itself, the genomic locus, the act of transcription, or any downstream smaller RNA by-products. X-inactivation is a striking example of a process controlled by LRGs acting at multiple levels and through diverse mechanisms. Our aim is to explore LRG’s versatility to understand whether a given LRG could play multiple functions in various developmental and cellular contexts, and how the underlying mechanisms may vary accordingly.
Publications
2024
- Morey C., Rougeulle C. & Ouimette JF. Unleashing XIST from X-chromosome inactivation. Current Opinion in Cell Biology, 2024 November
- Alfeghaly C., Castel G., Cazottes E., Moscatelli M., Moinard E., Casanova M., Boni J., Mahadik K., Lammers J., Freour T. , Chauviere L.,Piqueras C., Boers R., Boers J., Gribnau J.,David L., Ouimette JF. & Rougeulle C. XIST dampens X chromosome activity in a SPEN-dependent manner during early human development. Natural Structural and Molecular Biology, 2024 June
- Altered X-chromosome inactivation predisposes to autoimmunity . Science Advances, 2024 May
2023
- Rosspopoff O, Cazottes E, Huret C, Loda A, Collier A, Casanova M, Rugg-Gunn P, Heard E, Ouimette JF, Rougeulle C. Species-specific regulation of XIST by the JPX/FTX orthologs . Nucleic Acids Research, 2023 Feb
2022
- Mahadik K, Rougeulle C. Study of X Chromosome Activity Status in Human Naive Pluripotent Stem Cells Using RNA-FISH Methods Mol Biol. 2022;2416:239-255
2021
- Heard E, Rougeulle C. Digging into X chromosome inactivation. Science. 2021 Nov 19;374(6570):942-943.
- Moscatelli M., Rougeulle C. Dernières nouvelles du chromosome X: des principes généraux nuancés. Médecine/Sciences, 37 2 (2021) 152-158
2020
- Ouimette JF, Rougeulle C. Many XCI-ting routes to reach the eXACT dose. Nat Cell Biol. 2020 Dec;22(12):1397-1398
- Cazottes E., Rougeulle C. Straight to the X: Modeling Human X Chromosome Inactivation in hESCs by FGF Signal Blockade.Cell Stem Cell. 2020 Sep 3;27(3):352-353
- Patrat C, Ouimette JF, Rougeulle C. X chromosome inactivation in human development. Development 2020 Jan 3;147(1).
2019
- Casanova M, Moscatelli M, Chauvière LÉ, Huret C, Samson J, Liyakat Ali TM, Rosspopoff O, Rougeulle C. A primate-specific retroviral enhancer wires the XACT lncRNA into the core pluripotency network in humans. Nat Commun. 2019 Dec 11;10(1):5652.
2018
- Furlan G, Gutierrez Hernandez N, Huret C, Galupa R, van Bemmel JG, Romito A, Heard E, Morey C, Rougeulle C. The Ftx Noncoding Locus Controls X Chromosome Inactivation Independently of Its RNA Products. Mol Cell. 2018 Apr 20. pii: S1097-2765(18)30227-2.
2017
- Vallot C., Patrat C., Collier AJ., Huret C., Casanova M., Liyakat Ali TM., Tosolini M., Frydman N., Heard E., Rugg-Gunn P.J.and Rougeulle C. Competing action of XIST and XACT noncoding RNAs in the control of X chromosome activity during human early development. Cell Stem Cell. 2017 Jan 5;20(1):102-111.
2016
- Vallot C., Ouimette JF., Rougeulle C. Establishment of X chromosome inactivation and epigenomic features of the inactive X depend on cellular contexts. Bioessays. 2016 Sep;38(9):869-80
- Ouimette JF, Rougeulle C. How Many Non-coding RNAs Does It Take to Compensate Male/Female Genetic Imbalance? Adv Exp Med Biol. 2016;886:33-49
2015
- Vallot C*, Ouimette JF*, Makhlouf M, Féraud O, Pontis J, Côme J, Martinat C, Bennaceur-Griscelli A, Lalande M, Rougeulle C. Erosion of X Chromosome Inactivation in Human Pluripotent Cells Initiates with XACT Coating and Depends on a Specific Heterochromatin Landscape.Cell Stem Cell. 2015 May 7;16(5):533-46.
2014
- Makhlouf M.*, Ouimette JF.*, Oldfield A.*, Navarro P., Neuillet D. and Rougeulle C. A prominent and conserved role for YY1 in Xist transcriptional activation. Nat Commun. 2014 Sep 11;5:4878.
2013
- Häfner S. and Rougeulle C. La matière noire du génome. Pour la Science, 81: 58-63.
- Vallot C., Huret C., Lesecque Y., Resch A., Oudrhiri N., Bennaceur-Griscelli A., Duret L. and Rougeulle C. XACT, a long non-coding transcript coating the active X chromosome in human pluripotent cells. Nat Genet. 2013 Mar;45(3):239-41.
2012
- Vallot C. and Rougeulle C. Epigenetic stability of human pluripotent stem cells. « Epigenomics: From Chromatin Biology to Therapeutics » 118-133.
2011
- Romito A. and Rougeulle C. Origin and evolution of the long non-coding genes in the X-inactivation center. Biochimie. 2011 Nov;93(11):1935-42.
- Makhlouf M. and Rougeulle C. Linking X Chromosome Inactivation to Pluripotency: Necessity or Fate? Trends Mol Med. 2011 Jun;17(6):329-36.
2010
- Navarro P., Oldfield A., Legoupi J., Festuccia N., Dubois A., Attia M., Schoorlemmer J., Rougeulle C., Chambers I. and Avner P. Molecular coupling of Tsix regulation and pluripotency. Nature 2010 Nov 18;468(7322):457-60.
- Mitjavila-Garcia M., Bonnet M.L., Yates F., Haddad R., Oudrhiri N., Féraud O., Magniez A., Vallot C., Makhlouf M., Rougeulle C., Bennaceur-Griscelli A. and Turhan A.G. Partial reversal of the methylation pattern of X-linked gene HUMARA during hematopoietic differentiation of human embryonic stem cells. J Mol Cell Biol. 2010 Oct;2(5):291-8.
2009
- Navarro P., Chantalat S., Foglio M., Chureau C., Vigneau S., Clerc P. Avner P. and Rougeulle C. A role for non-coding Tsix transcription in partitioning chromatin domains within the mouse X-inactivation centre. Epigenetics Chromatin. 2009 Jul 20;2(1):8.
Members

Jeanne Brouillet
Bioinformatic engineer (CDD CNRS ERC XCycle)
brouillet.jeanne@parisepigenetics.com
+33(0)157278930
READ MORE
Romina Facchinello
PhD student (Université Paris Cité)
romina.facchinello@etu.u-paris.fr
+33 (0)157278930
READ MORE
Christophe Huret
Lab Manager | Vectorology platform manager (Université Paris Cité)
+33(0)157278929
READ MORE
Céline Morey
Senior researcher (INSERM)
celine.morey@univ-paris-diderot.fr
+33(0)157278929
READ MORE
Jean-Francois Ouimette
Assistant Professor (Université Paris Cité)
jean-francois.ouimette@u-paris.fr
+33(0)157278929
READ MOREAlumni
Megha Balachandra – Master 2 student
Naïs Boistol – BTS student
Juliette Boni – Master 1 student
Charlie Bouchu – Master 1 student
Syrlène Bouabdallah – Master 2 student
Miguel Casanova – Post-Doc
Léo Carrillo – PhD student
Gaël Castel – Bionformatic engineer
Emmanuel Cazottes – PhD student / transitional Post Doc
Louis Chauvière – PhD student
Solène Coste – Master 2 student
Arnaud Divol – Master 2 student
Giulia Furlan – PhD student / transitional Post Doc
Angélique Galvani – Lecturer
Elena Gomez Garcia – Licence 2 Erasmus student
Nancy Gutierrez Hernandez – Master 1 student
Sophia Häfner – PhD student
Margot Hully – Master 1 ENS student
Yoann Kovacs – Master 2 student
Ibrahim Lafi – Master 1 Polytechnique student
Alexia Le Barch – Master 1 Polytechnique Student
Tharvesh Moideen Liyakat Ali – Bioinformatic engineer
Charlie London – Engineer
Kasturi Mahadik – Post Doc
Mélanie Makhlouf – PhD student / Post Doc
Madeleine Moscatelli – PhD student / transitional Post Doc
Damien Neuillet – Assistant engineer
Andrew Oldfield – PhD student
Antonio Romito – Post Doc
Olga Rosspopoff – PhD student / transitional Post Doc
Julia Samson – Master 2 student
Samantha Sheng – Post Doc
Matteo Tossolini – Master 2 student
Céline Vallot – researcher CNRS
Valentina Varriale – Master 1 Polytechnique student
Contact
Claire ROUGEULLE, PhD, HDR, DR1 – CNRS
CNRS – Epigenetics and Cell Fate – UMR7216
Université Paris Cité
Bat Lamarck, 4ème étage. Case courrier 7042
35, rue Hélène BRION
75205 Paris Cedex 13
Tél : 33 (0)1 5727 8924
Fax : 33 (0)1 5727 8911
Email : claire.rougeulle@u-paris.fr
Related content
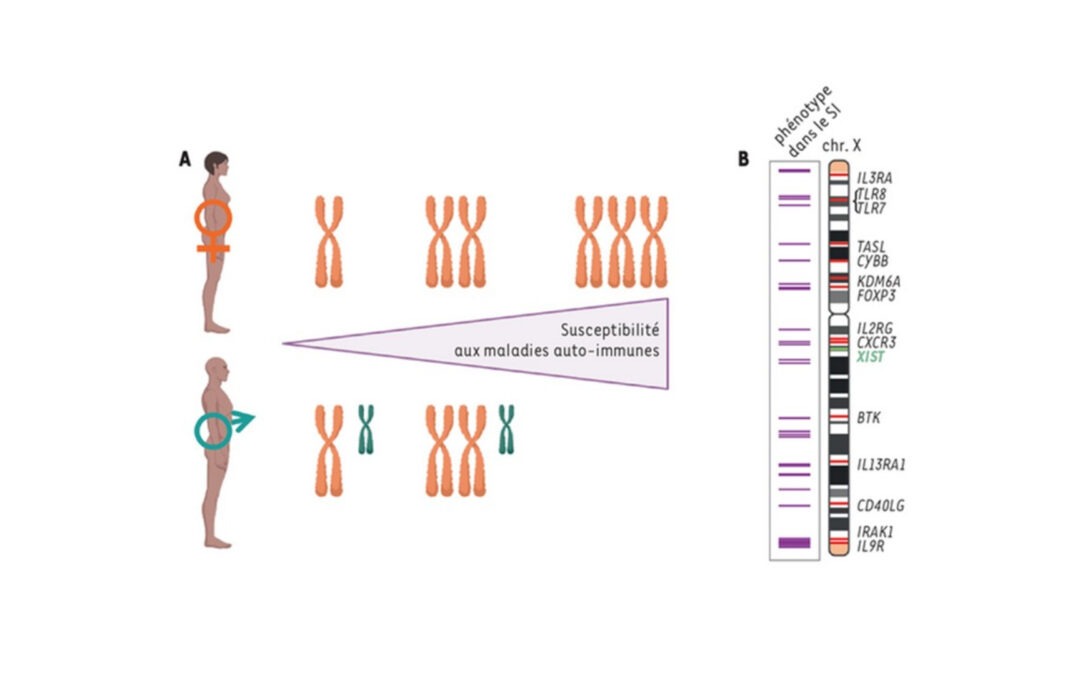
New review: X chromosome regulation and female functional specificities: Are two Xs better than one?
What if the presence of two X chromosomes confers functional specificities on female cells and contributes to the different susceptibilites of men and women to certain diseases? One of the X chromosomes is randomly silenced in each female cell from the embryonic...
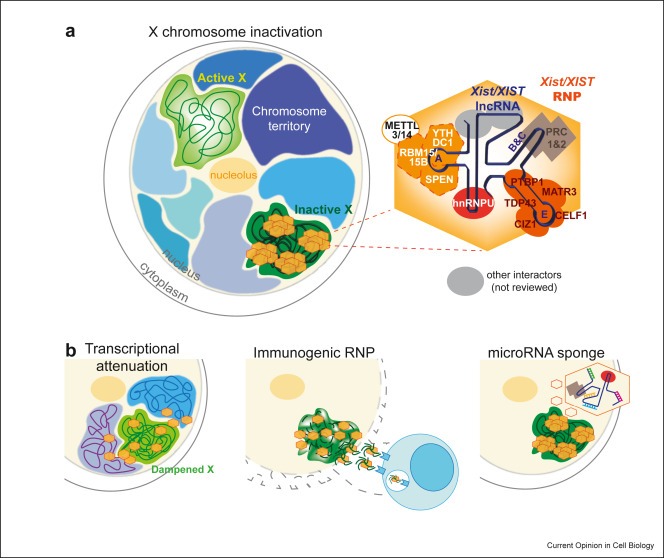
New review: Unleashing XIST from X-chromosome inactivation
The discovery that long noncoding RNAs (lncRNAs) are the most abundant gene class in the human genome has sparked significant research, particularly on Xist, a key RNA involved in X-chromosome inactivation. Recent studies have expanded our understanding of Xist's...

Congratulations Dr Carrillo
Congratulations to Léo who defended his thesis work on " Exploring the link between X chromosome inactivation and the development of human extraembryonic tissues " . © Rougeulle team À lire aussi
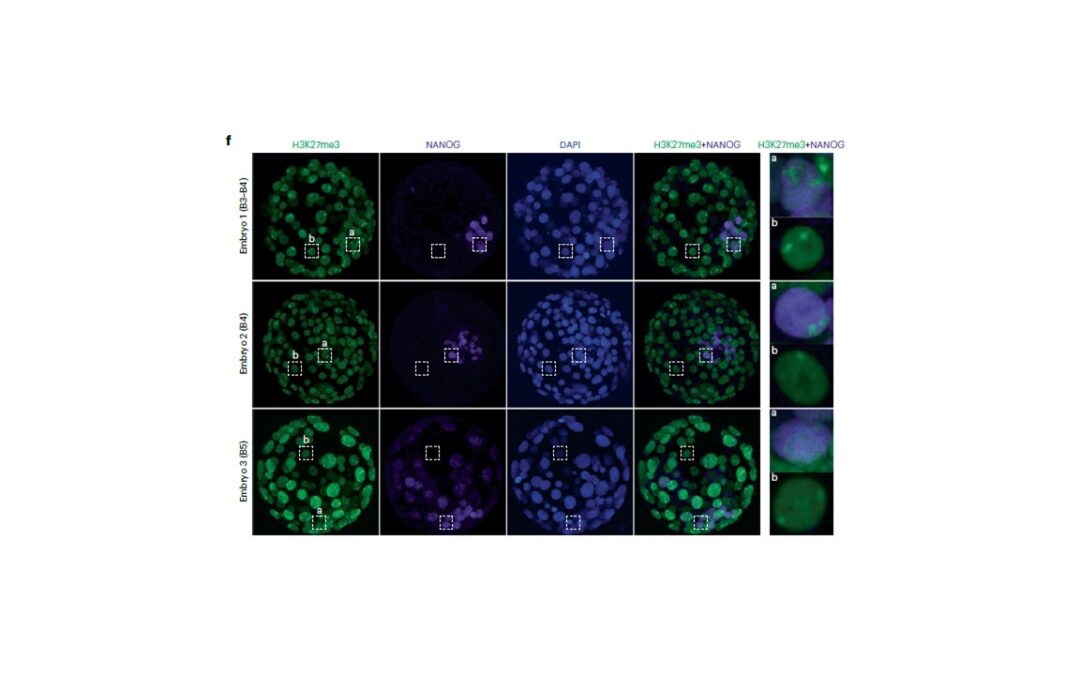
Article: XIST dampens X chromosome activity in a SPEN-dependent manner during early human development
Our latest work carried out by Alfeghaly C. et al. elucidating the mechanism of dampening of X-linked gene expression during pre-implantation development. To discover the role of the long non-coding RNA XIST and its particular partner SPEN click on the link: Nature...








