Polo group
Epigenome integrity
Sophie Polo’s group studies chromatin plasticity in response to DNA damage in mammalian cells. Research in her lab focuses on identifying the molecular players that control histone dynamics, alterations in chromatin marks and higher-order chromatin structure and function in response to genotoxic stress. Reciprocally, they investigate the impact of epigenome alterations on genome stability in physiological and pathological contexts. By combining proteomics with cutting-edge imaging, they are interested in understanding how genome and epigenome maintenance are coordinated.
Epigenome integrity group (April 2025)
From left to right:
Back: Sophie Polo, Annabelle Shaw, Audrey Chansard, Julia Roche Dupuy, Giulia Giacomini, Delphine Burlet.
Front: Sandra Piquet, Juliette Dabin, Margherita Mori, Eliane Petit.
In this context, we are interested in understanding how the information conveyed by chromatin structure is preserved when challenged by genotoxic stress and how genome and epigenome maintenance are coordinated in physiological conditions and during tumorigenesis. For this, we investigate DNA damage-induced alterations at distinct levels of chromatin organization in mammalian cells, from histone proteins up to higher-order chromatin structure, and we explore the underlying mechanisms. Reciprocally, we dissect the consequences of epigenome alterations, including histone oncomutations, on genome maintenance.
Our experimental approaches combine molecular biology and proteomics with advanced microscopy techniques to investigate chromatin dynamics in response to genotoxic stress in cultured mammalian cells. They include:
- the local induction of DNA lesions through micro-irradiation or the CRISPR/Cas9 technology,
- in vivo tracking of histone proteins using the SNAP-tag technology,
- the dynamic profiling of epigenetics marks – DNA and histone modifications – on repair patches.
Our research projects:
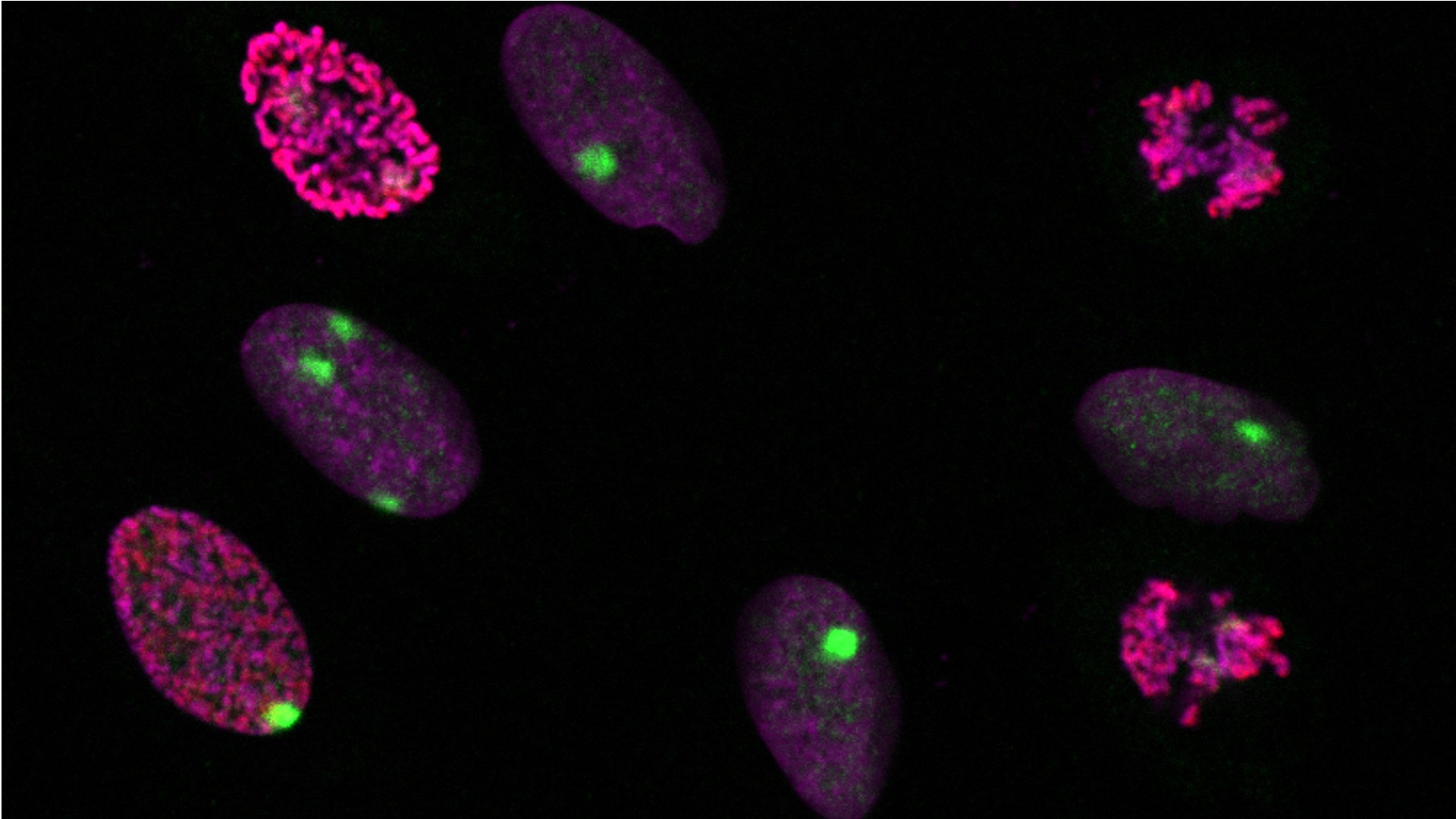
Dynamic profiling of DNA and histone modifications at repair sites
Description
To determine whether and how epigenetic states are stably inherited through DNA repair, we profile the epigenome of UV damage repair patches in mammalian cells, focusing on histone post-translational modifications and DNA methylation.
To characterize dynamic changes in the histone modification landscape during the repair of UV lesions in human cells in a comprehensive and unbiased manner, we designed two complementary proteomic approaches based on (1) proximity biotinylation of proteins at repair sites and on (2) capturing sites of repair synthesis, combined with mass spectrometry analyses. In parallel, we take a candidate approach focusing on mitotic histone phosphorylations at UV damage sites by imaging. We then employ functional studies to decipher the mechanisms underlying histone modification alterations during UV damage repair and their consequences on cell fate.
To investigate DNA methylation maintenance at sites of UV damage repair, we combine imaging and high-throughput sequencing approaches on nascent repaired DNA. We aim at identifying the molecular players involved, and potential cross-talks with histone dynamics.
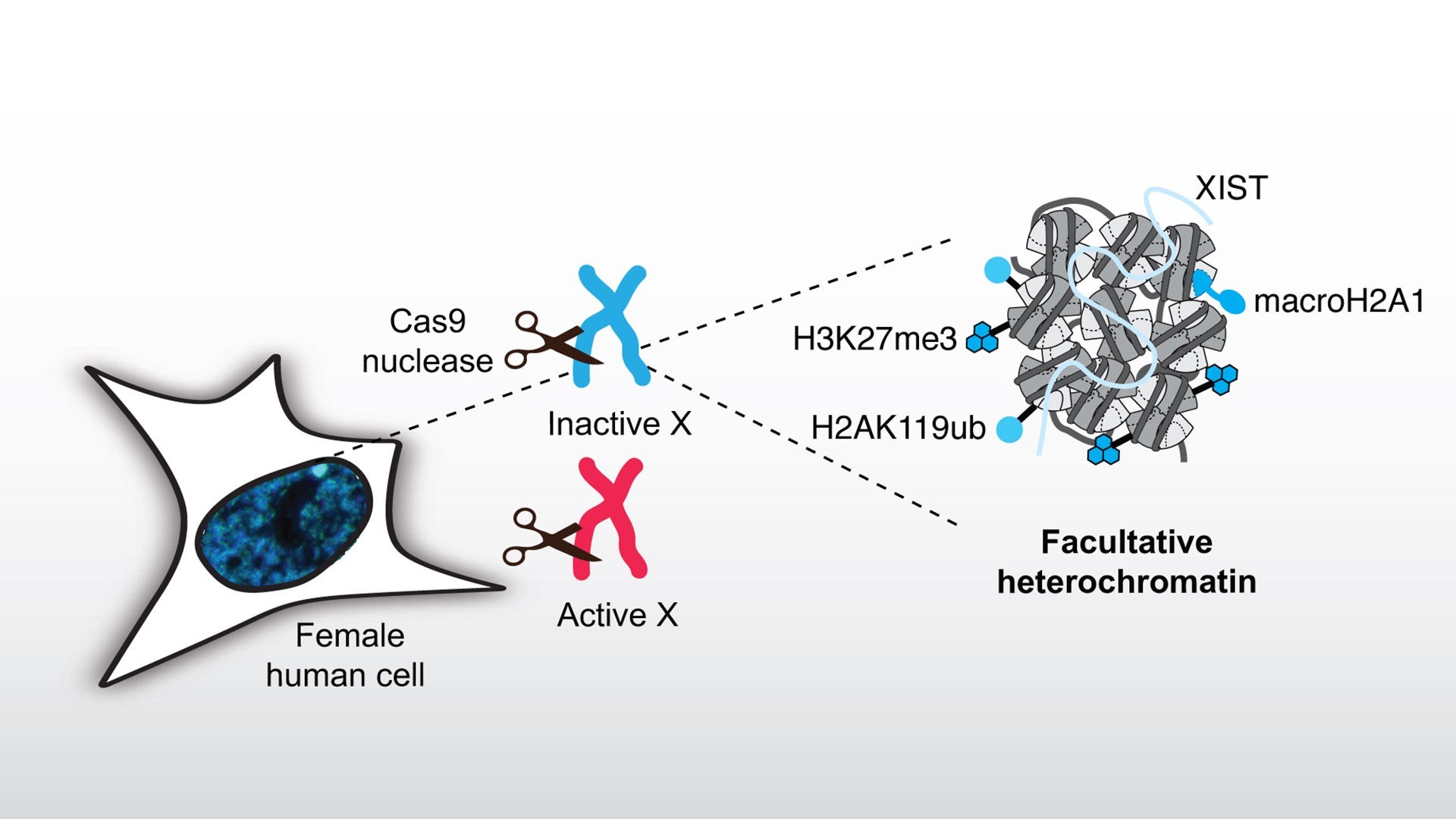
Exploring the response to DNA damage in facultative heterochromatin
Description
Compact heterochromatin domains represent challenging environments for DNA damage repair as shown by their high mutation rates. To study the response of facultative heterochromatin to DNA damage, we focus on the inactive X chromosome (Xi) that is silenced by facultative heterochromatin formation during embryonic development in female mammals.
We induce sequence-specific DNA double-strand breaks (DSBs) with Cas9 nuclease in X chromosomes and examine how the facultative heterochromatin landscape governs DSB repair pathway choice. We also test the hypothesis that DSB induction can guide the choice of the X chromosome to be inactivated in embryonic stem cells. Reciprocally, we analyze the impact of the DSB repair response on the maintenance/alteration of Xi chromatin marks and explore their functional outcome on X-linked gene silencing. In particular, we focus on alterations of the facultative heterochromatin mark H3K27me3 in response to DSBs and the functional relevance of fine-tuning this histone mark for genome stability.
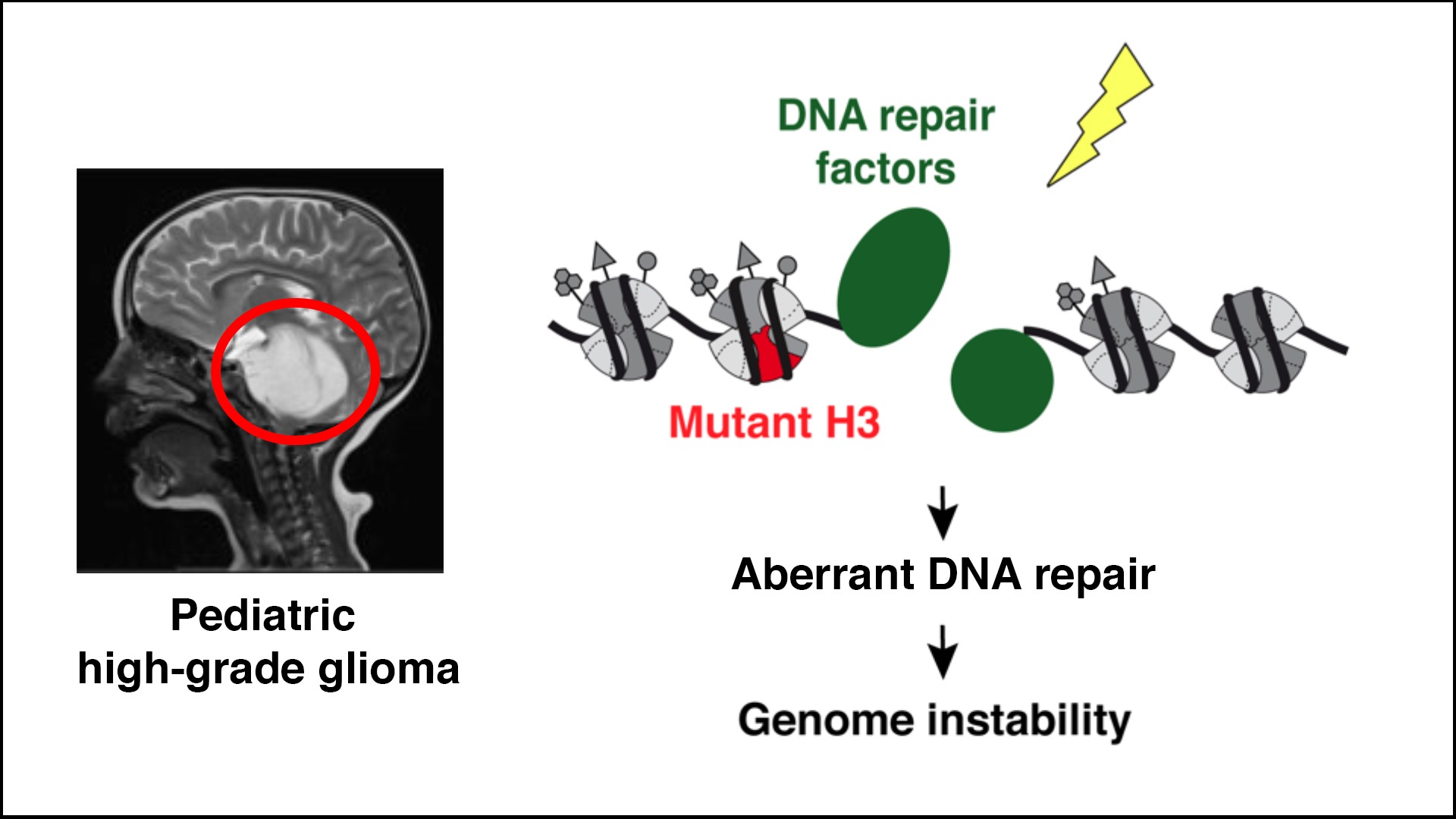
Dissecting the impact of histone oncomutations on genome maintenance
Description
Point mutations in histone H3 variants are driver events in several types of cancers including pediatric high-grade glioma (pHGG), a deadly and currently incurable disease. To open up new therapeutic options, we test the hypothesis that glioma-associated mutations in H3 histones (K27M, G34R/V) may trigger genome instability by altering DNA repair. Aberrant DNA repair may confer vulnerabilities that can be targeted to counteract cancer cell growth.
We combine genomics, proteomics and imaging approaches in pHGG tumors and cellular models with in vivo pre-clinical studies to (1) dissect and exploit the dysfunction of DNA repair enzymes in H3 mutant cells, (2) characterize the chromatin-based mechanisms connecting mutant histones with aberrant DNA repair, and (3) compare the response to DNA damage of H3.1 and H3.3 oncomutations.
Publications
2026
- Plessier A.*, Chansard A.*, Petit E.*, Novion Ducassou J., Couté Y., and Polo S.E.Proteomic profiling of UV damage repair patches uncovers histone chaperones with central functions in chromatin repair. Nat Commun, in press, 2026. *: equal contribution
2025
- Ferrand J.*, Dabin J.*, Chevallier O., Kane-Charvin M., Kupai A., Hrit J., Rothbart S., and Polo S.E. Mitotic chromatin marking drives the segregation of DNA damage. Nat Commun, 16: 746, 2025. *: equal contribution
2024
- Dabin J.*, Giacomini G.*, Petit E.*, and Polo S.E. New facets in the chromatin-based regulation of genome maintenance. DNA Repair, 140: 103702, 2024. *: equal contribution
- Saredi G., Carelli F.N., Rolland S.*, Furlan G.*, Piquet S.*, Appert A.*, Sanchez-Pulido L., Price J.L., Alcon P., Lampersberger L., Déclais A-C., Ramakrishna N.B., Toth R., Macartney T., Alabert C., Ponting C.P., Polo S.E., Miska E.A., Gartner A., Ahringer J., and Rouse J. The histone binding capacity of SPT2 controls chromatin structure and function in Metazoa. Nat Struct Mol Biol, 2024, doi : 10.1038/s41594-023-01204-3. *: equal contribution
- Giacomini G., Piquet S., Chevallier O., Dabin J., Bai S-K., Kim B., Siddaway R., Raught B., Coyaud E., Shan C-M., Reid R.J.D., Toda T., Rothstein R., Barra V., Wilhelm T., Hamadat S., Bertin C., Crane A., Dubois F., Bandopadhayay P., Beroukhim R., Naim V., Jia S., Hawkins C., Rondinelli B.* and Polo S.E.* Aberrant DNA repair reveals a vulnerability in histone H3.3-mutant brain tumors. Nucleic Acids Res, 2024. doi: 10.1093/nar/gkad1257. *: co-corresponding authors
2023
- Dabin J.*, Mori M.*, Polo S.E. The DNA damage response in the chromatin context: a coordinated process. Current Opin Cell Biol, 82: 102176, 2023. *: equal contribution
2022
- Chansard A.*, Pobega E.*, Caron P., Polo S.E. Imaging the response to DNA damage in heterochromatin domains. Front Cell Dev Biol, 10: 920267, 2022. *: equal contribution
2021
- Caron P.*, Pobega E.*, Polo S.E. DNA double-strand break repair: All roads lead to heterochROMAtin marks. Front Genet, 12: 730696, 2021. *: equal contribution
- Bouvier D., Ferrand J., Chevallier O., Paulsen M.T., Ljungman M., Polo SE. Dissecting regulatory pathways for transcription recovery following DNA damage reveals a non-canonical function of the histone chaperone HIRA. Nat Commun, 12: 3835, 2021.
- Olley G., Madapura P., Grimes G., Piquet S., Polo S.E., FitzPatrick D., Bickmore W., Boumendil C. Cornelia-de Lange syndrome-associated mutations cause a DNA damage signalling and repair defect. Nat Commun, 12: 3127, 2021.
- Caron P., Pobega E., Polo S.E. A molecular Rosetta Stone to decipher the impact of chromatin features on the repair of Cas9-mediated DNA double-strand breaks. Mol Cell, 81:2059-2060, 2021.
- Fortuny A., Chansard A., Caron P., Chevallier O., Leroy O., Renaud O., Polo S.E. Imaging the response to DNA damage in heterochromatin domains reveals core principles of heterochromatin maintenance. Nat Commun, 12: 2428, 2021.
- Ferrand J.*, Plessier A.*, Polo S.E. Control of the chromatin response to DNA damage: Histone proteins pull the strings. Semin Cell Dev Biol, 113:75-87, 2021. *: equal contribution
2020
- Ferrand J.*, Rondinelli B.*, Polo S.E. Histone variants: guardians of genome integrity. Cells, 9:2424, 2020. *: equal contribution
- Ahmad R, Lahuna O, Sidibe A, Daulat A, Zhang Q, Luka M, Guillaume JL, Gallet S, Guillonneau F, Hamroune J, Polo S, Prévot V, Delagrange P, Dam J, Jockers R. GPR50-Ctail cleavage and nuclear translocation: a new signal transduction mode for G protein-coupled receptors. Cell Mol Life Sci, 77: 5189-5205, 2020.
- Caron P., Polo S.E. Reshaping Chromatin Architecture around DNA Breaks. Trends Biochem Sci, 45: 177-179, 2020.
2018
- Piquet S., Le Parc F., Bai, S-K., Chevallier O., Adam S., Polo S.E. The histone chaperone FACT coordinates H2A.X-dependent signaling and repair of DNA damage. Mol Cell, 72: 888-901, 2018.
- Dabin J.*, Fortuny A.*, Piquet S, Polo S.E. Live imaging of parental histone variant dynamics in UVC-damaged chromatin. Methods Mol Biol, 1832: 243-253, 2018. *: equal contribution
- Fortuny A, Polo S.E. The response to DNA damage in heterochromatin domains. Chromosoma, 127: 291-300, 2018.
2017
- Fortuny A., Polo S.E. Genome and epigenome maintenance by keeping histone turnover in check. Mol Cell, 66: 3-4, 2017
- Dabin J., Polo S. E. Choreography of parental histones in damaged chromatin. Nucleus, 8: 255-260, 2017
- Polo S.E. Switching genes to silent mode near DNA double-strand breaks. EMBO Rep, 18:659-660, 2017.
2016
- Adam S.*, Dabin J.*, Chevallier O., Leroy O., Baldeyron C., Corpet A., Lomonte P., Renaud O., Almouzni G. and Polo S.E. Real-time tracking of parental histones reveals their contribution to chromatin integrity following DNA damage. Mol Cell, 64: 65-78, 2016. *: equal contribution. Featured article, also highlighted in the « Meet the author » section
- Dabin J.*, Fortuny A.* and Polo S.E. Epigenome maintenance in response to DNA damage. Mol Cell, 62: 712-727, 2016. *: equal contribution
2015
- Polo S.E.*, Almouzni G.* Chromatin plasticity in response to DNA damage: the legacy of the access-repair-restore model. DNA Repair, 36: 114-121, 2015. *: co-corresponding author
- Adam S.*, Dabin J.* and Polo S.E. Chromatin plasticity in response to DNA damage: the shape of things to come. DNA Repair, 32:120-126, 2015. *: equal contributio
- Adam S.*, Dabin J.*, Bai S-K. and Polo S.E. Imaging local deposition of newly synthesized histones in UVC-damaged chromatin. Methods Mol Biol, 1288-337-347, 2015. *: equal contributio
- Polo S.E. Reshaping chromatin after DNA damage: the choreography of histone proteins. J Mol Biol, 427:626-636, 2015.
2014
- Adam S., Polo S.E. Blurring the line between the DNA damage response and transcription: the importance of chromatin dynamics. Exp Cell Res, 329:148-153, 201
- Adam S., Polo S. E. and Almouzni G. How to restore chromatin structure and function in response to DNA damage – let the chaperones play. FEBS J, 281: 2315-2323, 2014
2013
- Adam S., Polo S. E.* and Almouzni G.* Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell, 155:94-106, 2013. *: co-corresponding author
- Polo S. E. Fine-tuning the p53 response to DNA damage: a new piece in the puzzle. Cell Cycle, 12: 1337-1338, 2013.
Members

Delphine Burlet
Post-doctoral fellow (INCa)
delphine.burlet-thomas@cnrs.fr
+33 1 57 27 89 32
READ MORE
Audrey Chansard
Research assistant | EPI2 Imaging Platform (CNRS)
audrey.chansard@univ-paris-diderot.fr
+33 1 57 27 89 81
READ MORE
Juliette Dabin
Post-doctoral fellow (MSCA)
juliette.dabin@univ-paris-diderot.fr
+33 1 57 27 89 81
READ MORE
Giulia Giacomini
Postdoctoral fellow (inIdEx FORMULA)
giulia.giacomini@etu.u-paris.fr
+33 1 57 27 89 32
READ MORE
Margherita Mori
Postdoctoral fellow (EUR GENE)
margherita.mori@etu.u-paris.fr
+33 1 57 27 89 32
READ MORE
Sandra Piquet
Research assistant & lab manager | EPI2 Imaging Platform manager (CNRS)
+33 1 57 27 89 81
READ MORE
Julia Roche Dupuy
PhD student (Univ. Paris Cité)
julia.roche-dupuy@etu.u-paris.fr
+33 1 57 27 89 32
READ MORE-
Past members
Salomé Adam
PhD student (Sept 2011-June 2015)
Postdoc (June 2015-Sept 2015)
After a highly succesful postdoc in Dan Durocher’s lab in Toronto (2015-2021), Salomé has moved to Cambridge UK to join AstraZeneca as a senior research scientist.
Siau-Kun Bai
Lab manager (Jan 2013-Mar 2014)
Siau is now research assistant in Ludger Johnannes’ laboratory at the Institut Curie, Paris.
Ines Beckerman
Visiting scientist (Sept-Dec 2024)
Ines went back to Dr. Manuel Muñoz laboratory at the University of Buenos Aires, Argentina to finish her PhD.
Zoé Begué
Master 1 student (Apr-June 2022)
Zoé has resumed her Master’s studies at Université Paris-Saclay.
Camille Boucher
Research assistant (July 2022-July 2023) as part of the Labex transverse program on intestinal organoids
Camille is now a research assistant in Beatrice Romagnolo’s lab at Institut Cochin, Paris
Déborah Bouvier
Research assistant (Jan 2016-Sept 2020)
Déborah is now a clinical research associate with IQVIA
Jeanne Brouillet
Master 1 student (June-Aug 2021)
Jeanne has resumed her Master’s studies at the AgroParisTech engineering school in Paris
Nolan Caile
Intern (Feb-Apr 2024)
Nolan has resumed his studies at Tufts University in Boston, USA
Pierre Caron
Post-doctoral fellow (Apr 2019-Dec 2021)
After another postdoc in Beatrice Eymin’s laboratory at the IAB in Grenoble, Pierre is now CNRS researcher in Joanna Timmins group at the Institute of Structural Biology in Grenoble
Odile Chevallier
Part time research assistant (Jan 2014-Feb 2023)
After 9 years of hard work in the team resulting in 4 published research papers and 2 in progress, Odile is now enjoying a well-deserved retirement.
Arnaud Debernardi
Master 1 student (Jan-Fev 2020)
Arnaud has resumed his Master’s studies at the University of Lyon
Marc El Hasbany
Master 1 student (Apr-July 2024)
After his internship on molecular dynamic simulations co-supervised by Leslie Regad (Univ. Paris Cité), Marc has resumed his Master’s studies at the University Paris Cité
Juliette Ferrand
Master’s student (Jan 2017-June 2017)
PhD student (Sept 2017-Sept 2021)
Post-doctoral fellow (Sept 2021-Dec 2022)
Juliette has moved to Copenhagen to join Niels Mailand lab as a postdoc.
Anna Fortuny
Master’s student (Jan 2015-June 2015)
PhD student (Sept 2015-Sept 2019)
Post-doctoral fellow (Sept 2019-Dec 2020)
Anna has moved back to Catalogna to join the R&D department of a biotech specialized in immunoassays.
Philippine Harou
Third year bachelor student (Nov-Dec 2023)
Philippine has resumed her studies at Université Paris Cité.
Matteo Kané-Charvin
Second year medical school student (June-Sept 2022)
Matteo has resumed his medical school studies at Université Paris Cité.
Florent Le Parc
Lab manager (Mar 2014-Dec 2015)
Florent has left academic science for training as an animal care taker.
François Martin
Intern (June-Aug 2023)
François has resumed his bio-engineering studies at Polytech Nice-Sophia.
Alexandre Plessier
Post-doctoral fellow (Apr 2019-Sept 2023)
Alex is now project manager at Université Paris Cité
Enrico Pobega
PhD student (Sept 2019-Dec 2023)
Enrico has moved back to the University of Trieste, Italy for his postdoctoral work.
Beatrice Rondinelli
Marie Curie postdoctoral fellow (Sept 2017- Sept 2018)
CNRS staff scientist (Oct 2018-April 2022)
Beatrice obtained an ATIP-avenir grant in 2021 allowing her to set up her own research group in May 2022 at the Gustave Roussy Institute
Hicham Ziaina
Master 1 student (Jan-Feb 2024)
Hicham has resumed his Master’s studies at the University Claude Bernard in Lyon.
Contact
Sophie Polo, PhD
Epigenetics and Cell Fate Centre
CNRS UMR7216 – Université Paris Cité
Lamarck B building, 4th floor, room 413
35, rue Hélène Brion
75205 Paris Cedex 13
Tel : 33 (0)1 57 27 89 81
Fax : 33 (0)1 57 27 89 11
Email : sophie.polo@u-paris.fr
We are always looking for enthusiastic and talented people to join our team. Please get in touch if you are interested in working with us.
Read more

Welcome to Léa
Léa joins the team as a research assistant. After completing a master's degree in virology, she worked in Strasbourg on grapevine viruses, then on characterizing mRNA degradation in plants at the Institute of Plant Molecular Biology (IBMP). In the Polo team, Léa will...

Sophie Polo receives an Impulscience® grant from the Fondation Bettencourt Schueller
Sophie Polo has been awarded an Impulscience® grant to fund a research project on the establishment and maintenance of the inactive X chromosome in response to DNA breaks. This is wonderful news for the lab ! We thank the Fondation Bettencourt Schueller for their...

Welcome to Léa, new engineer in the team!
Léa joins the lab as a research assistant. She holds a Master's degree in Molecular and Cellular Biology from Sorbonne University. She will contribute to investigate DNA methylation maintenance mechanisms in response to UV damage in mammalian cells. Léa Girard À lire...

Well done, Dr Mori!
Margherita successfully defended her PhD on DNA methylation maintenance in response to UV damage. Brava! Margherita and her thesis jury. From left to right: Sophie Polo, Sandra Duharcourt (on screen), Déborah Bourc'his, Margherita Mori, Nataliya Petryk, Jean Molinier,...


















