Ait-Si-Ali group
Epigenetic Dynamics & Cellular Differentiation

© Slimane Aït-Si-Ali, 2025
SYNOPSIS
A central epigenetic mechanism is the lysine methylation/demethylation of histones, as well as specific non-histone proteins. Lysine methylation on histones is central in imposing stable and heritable gene expression programs, being associated with both active and inactive domains of chromatin, depending on the residue that is targeted. Methylation of histone H3 on lysine 9 (H3K9) and lysine 27 (H3K27) being hallmarks of repressed chromatin. The enzymes controlling “histone” lysine methylation are called lysine methyltransferases (KMTs) and lysine demethylases (KDMs) and some can also modify non-histone substrates. They are highly specific for the targeted residue and, so far, around 50 KMTs and 30 KDMs have been described. We are particularly interested in KMTs specific for H3K9.
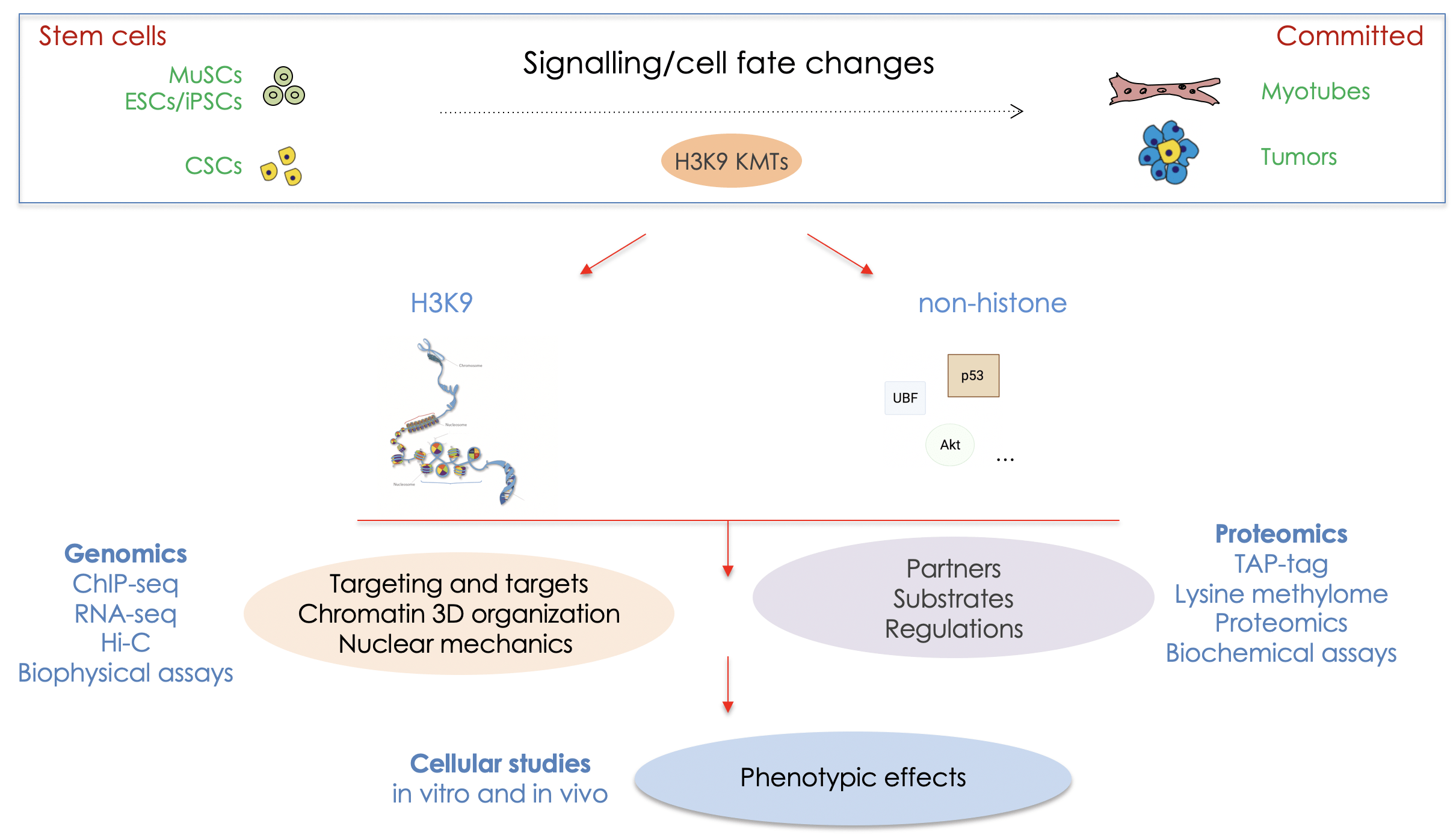
MAIN GOALS : CONTRIBUTION OF LYSINE METHYLATION BY THE H3K9 KMTs IN THE ESTABLISHMENT AND MAINTENANCE OF CELL IDENTITY
Unravelling the molecular mechanisms by which stem cells undergo cell fate decisions, especially differentiation, is one of the fundamental goals of modern medicine. The ability to modify a cell state is a fundamental issue that hold great promises for regenerative medicine.
Our group seeks to define the biochemical and molecular mechanisms that govern the normal silencing of genes during cell fate changes. We are interested in the role of lysine methylation, especially histone methylation of H3K9, as well as methylation of non-histone proteins by the H3K9 KMTs, in the regulation of cellular stemness and differentiation, in normal and pathological conditions such as cancer and dystrophies.
We study skeletal muscle stem cells (MuSCs), embryonic (ESCs) and induced pluripotent (iPSCs) and cancer stem (CSCs) cells. We dissect the composition, mechanisms, the kinetic of action of H3K9 KMTs and the crosstalk between them. Thus, the expected results will improve our knowledge on the mechanisms governing chromatin modifications during cellular fate changes, and have the potential to unravel new therapeutic strategies.
Our experimental approaches: to study KMTs at the molecular and cellular levels we combine gain- and loss-of-function strategies in ESCs, MuSCs and lung adenocarcinoma cancer stem cells. We use CRISPR-based genome editing, epigenomics (ChIP-seq, bulk and single-cell RNA-seq) and proteomics (TAP-tag/mass spectrometry), bioinformatics, biophysical studies of the nucleus by Atomic Force Microscopy (AFM) and microfluidic (in collaboration) approaches, cell biology studies and sophisticated imaging, automated wound healing and xenografts.
Publications
Selected recent publications
- Epigenetic control of nuclear mechanics and cellular migration via histone H3 lysine 9 methylation at Lamina-Associated Domains. Silvia Comunian, , , , , , , , , , , , ,
-
EMILIN1 emerges as a TGFβ/SETDB1-regulated secreted biomarker in Duchenne Muscular Dystrophy. bioRxiv 2025, Maeva Zamperoni, Laura Muraine, Alice Granados, Anne Bigot, Valentin Petit, Mona Bensalah, Jessica Ohana, Véronique Legros, Ekaterina Boyarchuk, Guillaume Chevreux, Véronique Joliot, Elisa Negroni, Maryline Moulin, Capucine Trollet, Slimane Ait-Si-Ali
DOI: 10.11.681604 - Automethylation of lysine methyltransferase SETDB1 on H3K9-like motifs regulates interactions with chromodomain proteins and controls its functions. bioRxiv, 2025. , , , , , , , ,
-
Genetic diseases of epigenetic machinery. Med Sci (Paris), 2024, Maud de Dieuleveult, Guillaume Velasco.
PMID: 39705562 -
Protocol for differential analysis of Trim71-associated protein complexes in mouse embryonic stem cells by mass spectrometry using Perseus. STAR Protoc, 2024, Cosson B, Rapone R, Del Maestro L, Ait-Si-Ali S.
PMID: 39116200 -
Setdb1 protects genome integrity in murine muscle stem cells to allow for regenerative myogenesis and inflammation. Dev Cell, 2024, Garcia P, Jarassier W, Brun C, Giordani L, Agostini F, Kung WH, Peccate C, Ravent J, Fall S, Petit V, Cheung TH, Ait-Si-Ali S, Le Grand F.
PMID: 38848717 - SETDB1 modulates the TGFβ response in Duchenne muscular dystrophy myotubes. Sci Adv, 2024, Granados A, Zamperoni M, Rapone R, Moulin M, Boyarchuk E, Bouyioukos C, Del Maestro L, Joliot V, Negroni E, Mohamed M, Piquet S, Bigot A, Le Grand F, Albini S, Ait-Si-Ali S.
PMID: 38691608 - The cytoplasmic fraction of the histone lysine methyltransferase Setdb1 is essential for embryonic stem cells. iScience, 2023, Rapone R, Del Maestro L, Bouyioukos C, Albini S, Cruz-Tapias P, Joliot V, Cosson B, Ait-Si-Ali S.
PMID: 37559904 - SETDB1 Fuels the Lung Cancer Phenotype by Modulating Epigenome, 3D Genome Organization and Chromatin Mechanical Properties. Nucleic Acids Research, 2022. Zakharova VV, Magnitov MD, Del Maestro L, Ulianov SV, Glentis A, Ulyanik B, Williart A, Karpukhina A, Demidov O, Joliot V, Vassetzky YS, Mège RM, Piel M, Razin SV & Ait-Si-Ali S.
PMID: 35474385 - Glucose controls co-translation of structurally related mRNAs via the mTOR and eIF2 pathways in human pancreatic beta cells. Front Endocrinol. 2022. Bulfoni M, Bouyioukos C, Zakaria A, Nigon F, Rapone R, Del Maestro L, Ait-Si-Ali S, Scharfmann R, Cosson B.
PMID : 35992129 - Paramecium Polycomb repressive complex 2 physically interacts with the small RNA-binding PIWI protein to repress transposable elements. Dev Cell. 2022. Del Maestro L, Ait-Si-Ali S,
PMID : 35429435
- Acute conversion of patient-derived Duchenne muscular dystrophy iPSC into myotubes reveals constitutive and inducible over-activation of TGFβ-dependent pro-fibrotic signaling. Skelet. Muscle 2020. Caputo L, Granados A, Lenzi J, Rosa A, Ait-Si-Ali S, Puri PL, Albini S.
PMID: 32359374 - The H3K9 Methylation Writer SETDB1 and Its Reader MPP8 Cooperate to Silence Satellite DNA Repeats in Mouse Embryonic Stem Cells. Genes, 2019. Cruz-Tapias P, Robin P, Pontis J, Del Maestro L, Ait-Si-Ali S.
PMID: 31557926 - Expression of the Major and Pro-Oncogenic H3K9 Lysine Methyltransferase SETDB1 in Non-Small Cell Lung Cancer. Cancers, 2019. Cruz-Tapias P, Zakharova V, Perez-Fernandez OM, Mantilla W, Ramírez-Clavijo S, Ait-Si-Ali S.
PMID: 31398867 - Canonical Wnt signalling regulates nuclear export of Setdb1 during skeletal muscle terminal differentiation. Cell Discovery, 2016. S Beyer, J Pontis, E Schirwis, V Battisti, A Rudolf, F Le Grand, S Ait-Si-Ali.
PMID: 27867535 - Unexpected antagonistic roles of the related histone H3 lysine 9 methyltarsferase G9a and G9a-Like Protein in proliferating but not in differentiating myoblasts. J Mol Biol, 2016. V Battisti, J Pontis, Boyarchuk E, Fritsch L, P Robin, S Ait-Si-Ali and V Joliot.
PMID: 27056598 - Sound of Silence: the properties and functions of repressive lysine methyltransferases. Nature Reviews Molecular Cell Biology, 2015. Mozzetta C, Boyarchuk E, Pontis J and Ait-Si-Ali S.
PMID: 26204160
Contact
CNRS – Epigenetics and Cell Fate – UMR7216
Université Paris Cité
Bâ Lamarck, 4ème étage. Case courrier 7042
35, rue Hélène BRION
75205 Paris Cedex 13
Tél : 33 (0)1 5727 8919
Fax : 33 (0)1 5727 8911
Email : slimane.aitsiali@u-paris.fr
Membres
Financements
Read more

Welcome to Léa
Léa joins the team as a research assistant. After completing a master's degree in virology, she worked in Strasbourg on grapevine viruses, then on characterizing mRNA degradation in plants at the Institute of Plant Molecular Biology (IBMP). In the Polo team, Léa will...

Sophie Polo receives an Impulscience® grant from the Fondation Bettencourt Schueller
Sophie Polo has been awarded an Impulscience® grant to fund a research project on the establishment and maintenance of the inactive X chromosome in response to DNA breaks. This is wonderful news for the lab ! We thank the Fondation Bettencourt Schueller for their...

Welcome to Léa, new engineer in the team!
Léa joins the lab as a research assistant. She holds a Master's degree in Molecular and Cellular Biology from Sorbonne University. She will contribute to investigate DNA methylation maintenance mechanisms in response to UV damage in mammalian cells. Léa Girard À lire...

Well done, Dr Mori!
Margherita successfully defended her PhD on DNA methylation maintenance in response to UV damage. Brava! Margherita and her thesis jury. From left to right: Sophie Polo, Sandra Duharcourt (on screen), Déborah Bourc'his, Margherita Mori, Nataliya Petryk, Jean Molinier,...