Defossez group
Dynamics and interpretation of DNA methylation in mammals

DEFOSSEZ team
Dr. Pierre-Antoine Defossez’s team is interested in the epigenetic mechanisms of regulation of gene expression in mammals.
Epigenetics : important in biology and human health
Epigenetics is the study of changes in gene function that do not involve changes to the underlying DNA sequence. These changes can include the attachment of chemical groups (such as methyl groups) to the DNA molecule or the histone proteins around which DNA is wrapped to form chromosomes. These modifications can turn genes “on” or “off” and affect how cells read the genes’ instructions.
Epigenetics is important because it helps explain how the environment can affect the way genes are expressed, and how these changes can be passed down from one generation to the next. For example, studies have shown that certain environmental factors, such as diet and stress, can lead to changes in the epigenetic marks on a person’s genes. These changes can then affect the person’s health and increase the risk of certain diseases.
Epigenetics is also important in the field of medicine because it could provide new targets for therapies. For example, drugs that target enzymes involved in adding or removing epigenetic marks could be used to treat cancer or other diseases caused by changes in gene expression. Additionally, understanding epigenetics could help researchers understand how to prevent certain diseases by identifying environmental factors that affect the epigenetic marks on a person’s genes.
DNA methylation : a paradigmatic epigenetic mark with key functions
DNA methylation is a process by which methyl groups are added to the DNA molecule. This process plays an important role in stem cells, as it helps to regulate the balance between self-renewal and differentiation. In stem cells, DNA methylation helps to silence genes that promote differentiation, allowing the stem cells to maintain their ability to divide and produce more stem cells. Additionally, DNA methylation is also important in the process of reprogramming adult cells into induced pluripotent stem cells, which have the ability to differentiate into any type of cell in the body.
In cancer, DNA methylation is often associated with the silencing of tumor suppressor genes, which are responsible for preventing the uncontrolled growth of cells. When these genes are silenced through DNA methylation, cancer cells can continue to divide and grow without any restriction. Additionally, cancer cells also exhibit global hypomethylation, meaning that the DNA of cancer cells is less methylated compared to normal cells. This global hypomethylation can contribute to the development of cancer by disrupting the regulation of many genes involved in cell growth and division. Drugs called DNA methyltransferase inhibitors are developed to target this process and used as cancer therapy.
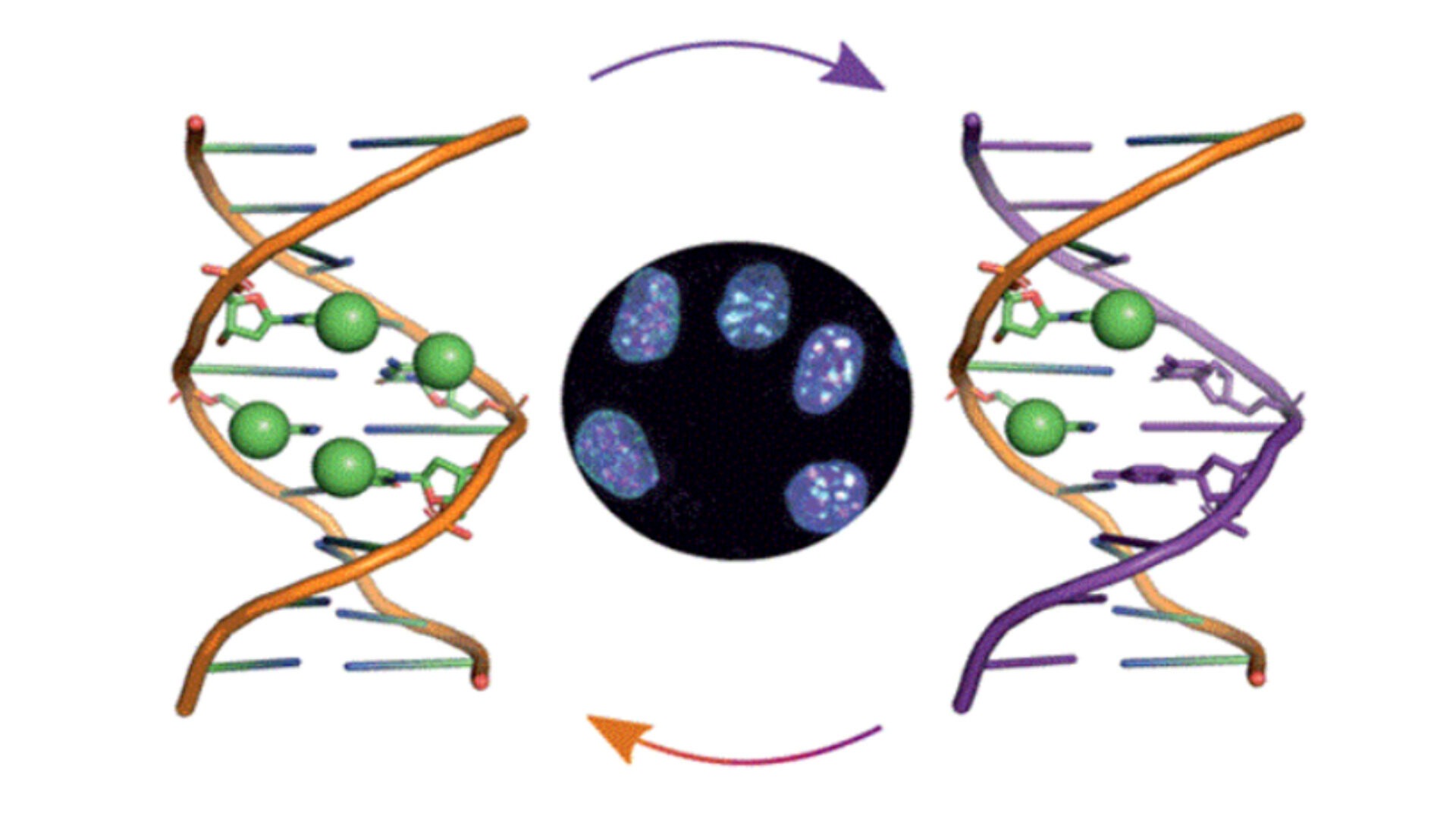
DNA methylation in stem cells
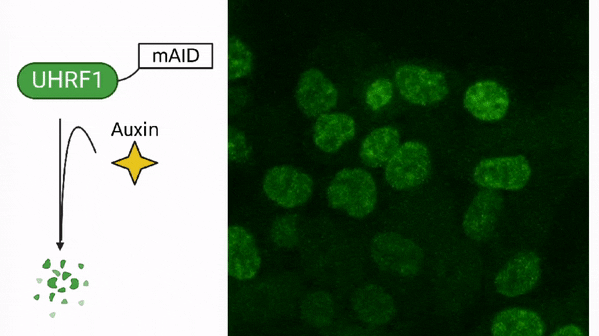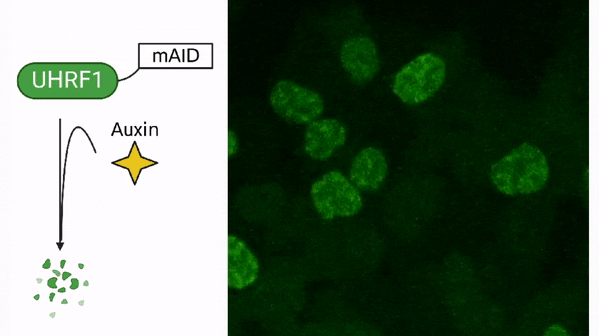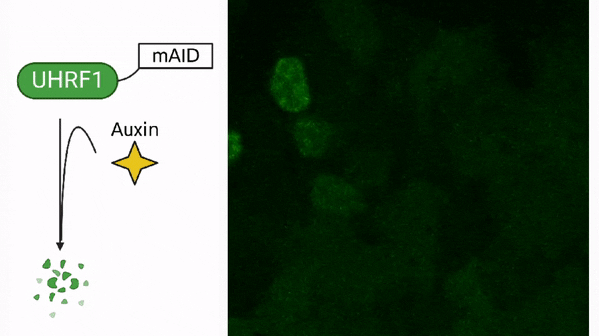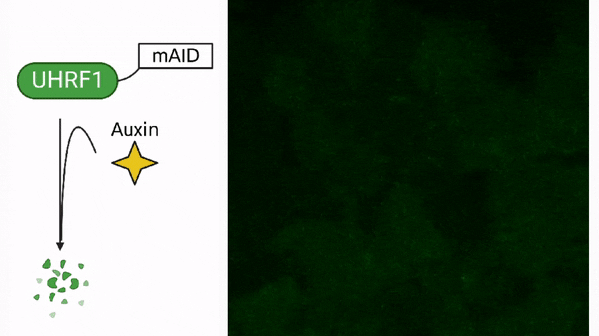
DNA methylation in cancer
Publications
Yamaguchi K, Chen X, Rodgers B, et al. Non-canonical functions of UHRF1 maintain DNA methylation homeostasis in cancer cells. Nat Commun 15, 2960 (2024). doi:10.1038/s41467-024-47314-4
Marthe Laisné, Sarah Benlamara, André Nicolas, et al. Cancer/Testis genes are predictive of breast tumor subtypes. Oncogene. 2024 May;43(18):1369-1385. doi: 10.1038/s41388-024-03002-7
Lanciano S, Philippe C, Sarkar A, et al. Comprehensive locus-specific L1 DNA methylation profiling reveals the epigenetic and transcriptional interplay between L1s and their integration sites. Cell Genomics. 2024 Jan 30:100498. doi: 10.1016/j.xgen.2024.100498
Chua BH, Zaal Anuar N, Ferry L, et al. E4F1 and ZNF148 are transcriptional activators of the -57A > C and wild-type TERT promoter. Genome Res. Published online November 2, 2023. doi:10.1101/gr.277724.123
Yakhou L, Azogui A, Gupta N, et al. A genetic screen identifies BEND3 as a regulator of bivalent gene expression and global DNA methylation. Nucleic Acids Res. Published online August 31, 2023:gkad719. doi:10.1093/nar/gkad719
Gupta N, Yakhou L, Albert JR, et al. A genome-wide screen reveals new regulators of the 2-cell-like cell state. Nat Struct Mol Biol. 2023;30(8):1105-1118. doi:10.1038/s41594-023-01038-z
Kikuchi, Amika, Hiroki Onoda, Kosuke Yamaguchi, Satomi Kori, Shun Matsuzawa, Yoshie Chiba, Shota Tanimoto, et al. “Structural Basis for Activation of DNMT1.” Nature Communications 13, no. 1 (November 21, 2022): 7130, doi: 10.1038/s41467-022-34779-4
Marchal, P.-A. Defossez, and B. Miotto, ‘Context-dependent CpG methylation directs cell-specific binding of transcription factor ZBTB38’, Epigenetics, pp. 1–22, Aug. 2022, doi: 10.1080/15592294.2022.2111135.
Yamaguchi, X. Chen, A. Oji, I. Hiratani, and P.-A. Defossez, ‘Large-Scale Chromatin Rearrangements in Cancer’, Cancers (Basel), vol. 14, no. 10, p. 2384, May 2022, doi: 10.3390/cancers14102384.
Kori S, Shibahashi Y, Ekimoto T, Nishiyama A, Yoshimi S, Yamaguchi K, Nagatoishi S, Ohta M, Tsumoto K, Nakanishi M, Defossez PA, Ikeguchi M, Arita K. Structure-based screening combined with computational and biochemical analyses identified the inhibitor targeting the binding of DNA Ligase 1 to UHRF1. Bioorg Med Chem. 2021 Dec 15;52:116500. doi: 10.1016/j.bmc.2021.116500. Epub 2021 Nov 10.
Petryk N, Reverón-Gómez N, González-Aguilera C, Dalby M, Andersson R, Groth A. Genome-wide and sister chromatid-resolved profiling of protein occupancy in replicated chromatin with ChOR-seq and SCAR-seq. Nat Protoc. 2021 Sep;16(9):4446-4493. doi: 10.1038/s41596-021-00585-3.
Blin M, Lacroix L, Petryk N, Jaszczyszyn Y, Chen CL, Hyrien O, Le Tallec B. DNA molecular combing-based replication fork directionality profiling. Nucleic Acids Res. 2021 Jul 9;49(12):e69. doi: 10.1093/nar/gkab219.
Petryk N, Bultmann S, Bartke T, Defossez PA. Staying true to yourself: mechanisms of DNA methylation maintenance in mammals. Nucleic Acids Res. 2021 Apr 6;49(6):3020-3032. doi: 10.1093/nar/gkaa1154.
Baubec T, Defossez PA. Reading DNA Modifications. J Mol Biol. 2020 Feb 8:S0022-2836(20)30096-6. doi: 10.1016/j.jmb.2020.02.001.
Cornett EM, Ferry L, Defossez PA, Rothbart SB. Lysine Methylation Regulators Moonlighting outside the Epigenome. Mol Cell. 2019 Sep 19;75(6):1092-1101. doi: 10.1016/j.molcel.2019.08.026.
Naciri I, Laisné M, Ferry L, Bourmaud M, Gupta N, Di Carlo S, Huna A, Martin N, Peduto L, Bernard D, Kirsh O, Defossez PA. Genetic screens reveal mechanisms for the transcriptional regulation of tissue-specific genes in normal cells and tumors. Nucleic Acids Research, 07 February 2019 gkz080, https://doi.org/10.1093/nar/gkz080
Kori S, Ferry L, Matano S, Jimenji T, Kodera N, Tsusaka T, Matsumura R, Oda T, Sato M, Dohmae N, Ando T, Shinkai Y, Defossez PA, Arita K. Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation. Structure. 2018 Dec 12. pii: S0969-2126(18)30455-6. doi: 10.1016/j.str.2018.11.012.
Laisné M, Gupta N, Kirsh O, Pradahan S, Defossez PA. « Mechanisms of DNA Methyltransferase Recruitment in Mammals ». Genes 2018, 9(12), 617; doi: 10.3390/genes9120617
Marchal C, de Dieuleveult M, Saint-Ruf C, Guinot N, Ferry L, Olalla Saad ST, Lazarini M, Defossez PA, Miotto B. Depletion of ZBTB38 potentiates the effects of DNA demethylating agents in cancer cells via CDKN1C mRNA up-regulation. Oncogenesis. 2018 Oct 11;7(10):82. doi: 10.1038/s41389-018-0092-0.
Ma X, Warnier M, Raynard C, Ferrand M, Kirsh O, Defossez PA, Martin N, Bernard D. « The nuclear receptor RXRA controls cellular senescence by regulating calcium signaling ». Aging Cell. 2018 Sep 14:e12831. doi: 10.1111/acel.12831.
Miotto B, Marchal C, Adelmant G, Guinot N, Xie P, Marto JA, Zhang L, Defossez PA. « Stabilization of the methyl-CpG binding protein ZBTB38 by the deubiquitinase USP9X limits the occurrence and toxicity of oxidative stress in human cells. » Nucleic Acids Res. 2018 Feb 27. doi: 10.1093/nar/gky149.
Ferry L, Fournier A, Tsusaka T, Adelmant G, Shimazu T, Matano S, Kirsh O, Amouroux R, Dohmae N, Suzuki T, Filion GJ, Deng W, de Dieuleveult M, Fritsch L, Kudithipudi S, Jeltsch A, Leonhardt H, Hajkova P, Marto JA, Arita K, Shinkai Y, Defossez PA. “A histone mimic within DNA Ligase 1 links DNA replication and DNA remethylation: a revised model for the maintenance of DNA methylation by UHRF1”. Molecular Cell , 2017 Aug 17;67(4):550-565.e5. doi: 10.1016/j.molcel.2017.07.012. Epub 2017 Aug 10.
Naciri , A. Roussel-Gervais , P.-A. Defossez. O. Kirsh. “Nouvelles fonctions d’une protéine liant l’ADN méthylé dans le cancer”, Médecine/Sciences (nouvelle), Volume 33, Numéro 8-9, Août–Septembre 2017. https://doi.org/10.1051/medsci/20173308009
Roussel-Gervais , I. Naciri , O. Kirsh , L. Kasprzyk , G. Velasco , G. Grillo , P. Dubus, P.-A. Defossez..“Loss of the methyl-CpG binding protein ZBTB4 alters the mitotic checkpoint, increases aneuploidy, and promotes tumorigenesis”, Cancer Research, 2016 Nov 4. pii: canres.1181.2016
Santolini M, Sakakibara I, Gauthier M, Ribas-Aulinas F, Takahashi H, Sawasaki T, Mouly V, Concordet JP, Defossez PA, Hakim V, Maire P. « MyoD reprogramming requires Six1 and Six4 homeoproteins: genome-wide cis-regulatory module analysis. » Nucleic Acids Res. 2016 Oct 14;44(18):8621-8640. Epub 2016 Jun 14.
Ferrand, O. Kirsh, A. Griveau, D. Vindrieux, N. Martin, P.-A. Defossez, and D. Bernard, “Screening of a kinase library reveals novel pro-senescence kinases and their common NF-κB-dependent transcriptional program,” Aging (Albany NY), vol. 7, no. 11, pp. 986–1003, Nov. 2015.
Miotto, M. Chibi, P. Xie, S. Koundrioukoff, H. Moolman-Smook, D. Pugh, M. Debatisse, F. He, L. Zhang, and P.-A. Defossez, “The RBBP6/ZBTB38/MCM10 axis regulates DNA replication and common fragile site stability,” Cell Rep, vol. 7, no. 2, pp. 575–587, Apr. 2014.
Contact
Pierre-Antoine DEFOSSEZ
pierre-antoine.defossez@cnrs.fr
Team members

Pierre-Antoine Defossez
Team Leader | Deputy Director
pierre-antoine.defossez@univ-paris-diderot.fr
READ MOREFunding




Read more

Welcome to Léa
Léa joins the team as a research assistant. After completing a master's degree in virology, she worked in Strasbourg on grapevine viruses, then on characterizing mRNA degradation in plants at the Institute of Plant Molecular Biology (IBMP). In the Polo team, Léa will...

Sophie Polo receives an Impulscience® grant from the Fondation Bettencourt Schueller
Sophie Polo has been awarded an Impulscience® grant to fund a research project on the establishment and maintenance of the inactive X chromosome in response to DNA breaks. This is wonderful news for the lab ! We thank the Fondation Bettencourt Schueller for their...

Welcome to Léa, new engineer in the team!
Léa joins the lab as a research assistant. She holds a Master's degree in Molecular and Cellular Biology from Sorbonne University. She will contribute to investigate DNA methylation maintenance mechanisms in response to UV damage in mammalian cells. Léa Girard À lire...

Well done, Dr Mori!
Margherita successfully defended her PhD on DNA methylation maintenance in response to UV damage. Brava! Margherita and her thesis jury. From left to right: Sophie Polo, Sandra Duharcourt (on screen), Déborah Bourc'his, Margherita Mori, Nataliya Petryk, Jean Molinier,...