Francastel group
DNA methylation and non-coding RNA in health and disease

group by Zoom
The 4 members of the Francastel team: Claire Francastel, DR2 INSERM; Florent Hubé, CRCN CNRS; Guillaume Velasco, MCU Université Paris Cité; Katia Boyarchuk, Engineer CNRS.
© EDC
The overall interest of our team is on the multiplicity and intricacy of the regulatory networks that control expression and maintenance of mammalian genomes, from local transcriptional and epigenetic control to global genome organization within the nuclear space. With the genomes of complex eukaryotes being pervasively transcribed but mostly made up of large amounts of sequences that do not carry information to make proteins, a challenging task is to document this huge non-coding transcriptional output and understand its functional significance in regulatory networks that govern normal cell fate. We are particularly focused on understanding whether and how perturbations to this non-coding production is a driving force is disease, and which are the mechanisms and factors that maintain their normal biogenesis and hence, genome and cell integrity. Over the past years, we focused on inherently non-coding genomic regions that represent a vast fraction of mammalian genomes, i.e. repeated sequences and introns, as paradigms to address these questions and as unconventional targets of (post)transcriptional and epigenetic perturbations that underpin human diseases.
The strategies of our team are both basic and patient-oriented, based on (i) the development of mouse models, (ii) the establishment of cohorts of patients in collaboration with physicians, (iii) a combination of complementary expertise in the study of gene expression, DNA methylation, non-coding RNA and their associated complexes, RNA splicing and processing and nuclear architecture, and (iv) capitalize on unique cellular models that we developed, including mouse models, ES cells and cells from patients, and the generation of high throughput data. Technical approaches include classical molecular biology, high throughput techniques for epigenome and transcriptome analysis, cellular imaging and in situ assays in the mouse.
Our main goals are to:
– document the non-coding transcriptional output from these regions in normal and physiopathological situations
– characterize the biogenesis of the produced transcripts, their processing, epigenetic regulation, sub-cellular localization and associated complexes
– understand if and how their deregulation represents a driving force in disease
– provide an integrated view of pan-genomic epigenetic, transcriptional and splicing defects in the context of pathological defects of the DNA methylation machinery.
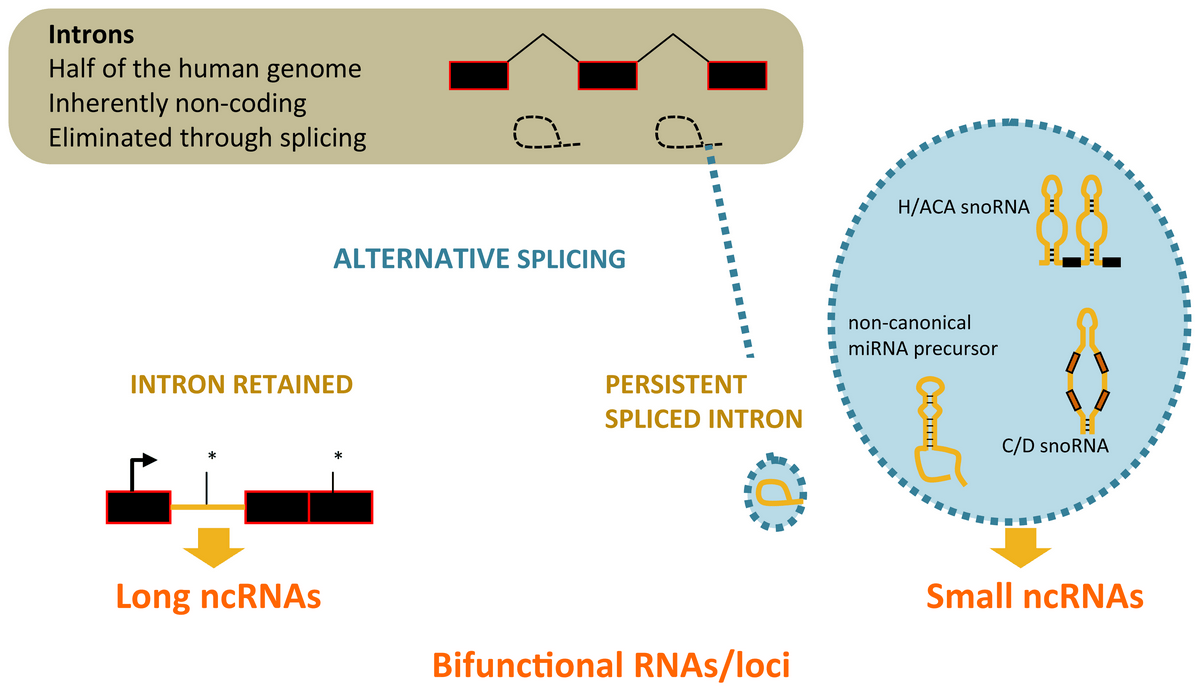
Alternative splicing of introns and the diversification of transcriptional output
A paradigm to test the functionality of non-protein-coding genomic regions
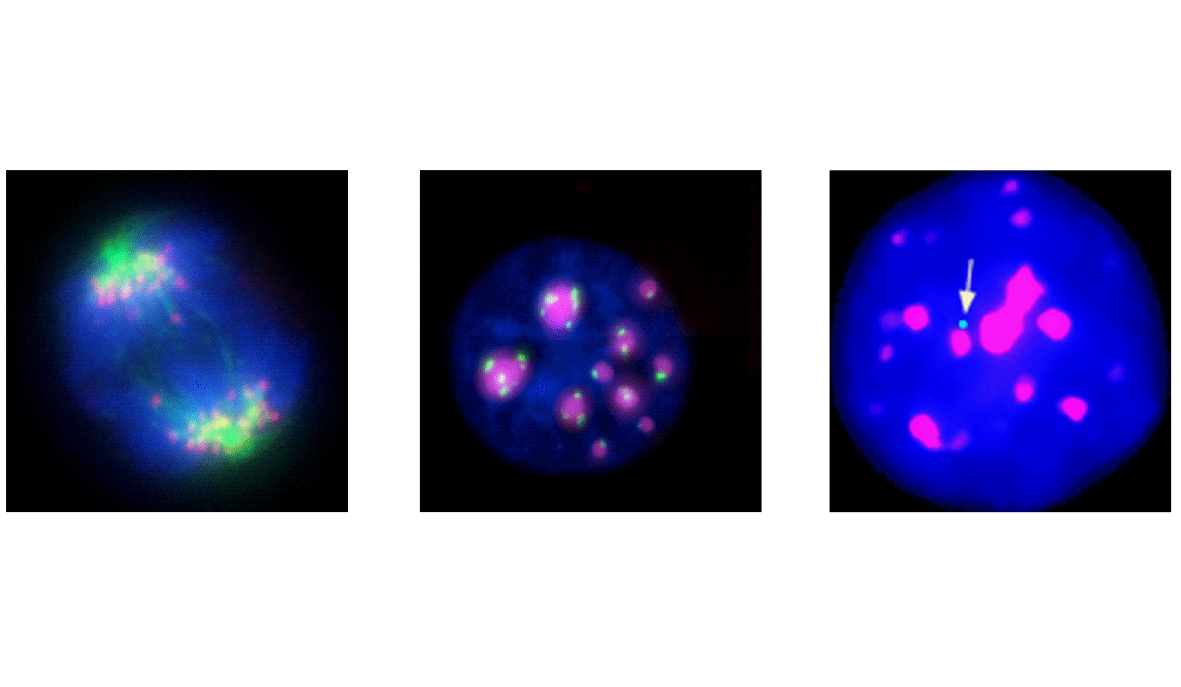
Centromeric repeats transcription
A paradigm to link transcription of DNA repeats to global molecular and cellular effects
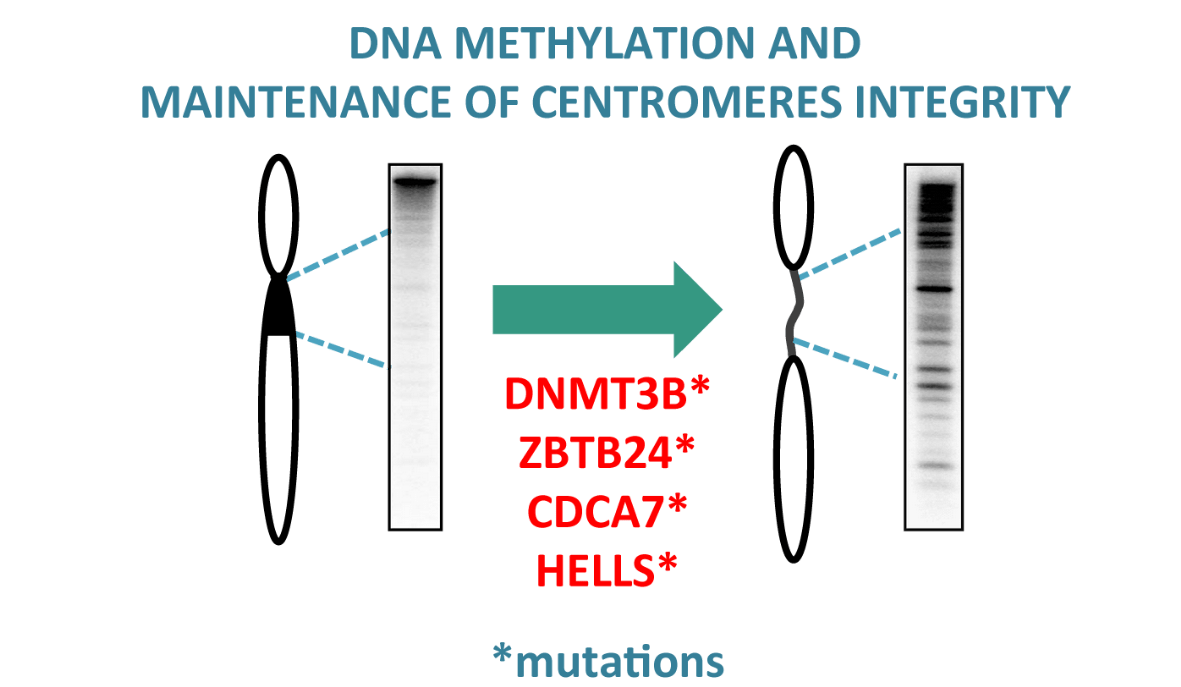
DNA methylation and genome maintenance
When studying a rare disease sheds new light on the field of DNA methylation
Publications
Contact
Membres
Read more

Welcome to Orlane
Today, we welcome a new member to the team: Orlane. She's a student from the first year of the Brevet de Technicien Supérieur Biotechnologies en Recherche et Production (Two-year technical degree - Biotechnologies in Research and Production).Orlane will be working...

4th year PhD fellowships for Anaëlle
Congratulations to Anaëlle Azogui for obtaining a 4th year PhD fellowships from the "Fondation pour la recherche sur le cancer" (Fondation ARC). Read more

Congratulations to the Defossez team for their article published in Nature Communication
Congratulations to the Defossez team for their article “DNA methylation protects cancer cells against senescence” by Chen, Yamaguchi, et al., which has been accepted in Nature Communication. Read it here: doi:doi: https://doi.org/10.1101/2024.08.23.609297...

Welcome to Delphine, new post-doc in the team!
Delphine joins the lab as a post-doctoral fellow. She completed her PhD at the Centre de Recherche en Cancérologie de Lyon (CRCL) under the supervision of Virginie Petrilli. Her doctoral work focused on the pro-inflammatory protein NLRP3, for which she highlighted a...
