EpiG : Functional epigenomics
The Functional Epigenomics platform’s objectives are to provide equipment and to train staff in its use.
It offers services for epigenome analysis and participates in the development of new techniques according to the evolution of the field.
The Functional Epigenomics platform is open to the academic and industrial scientific community.
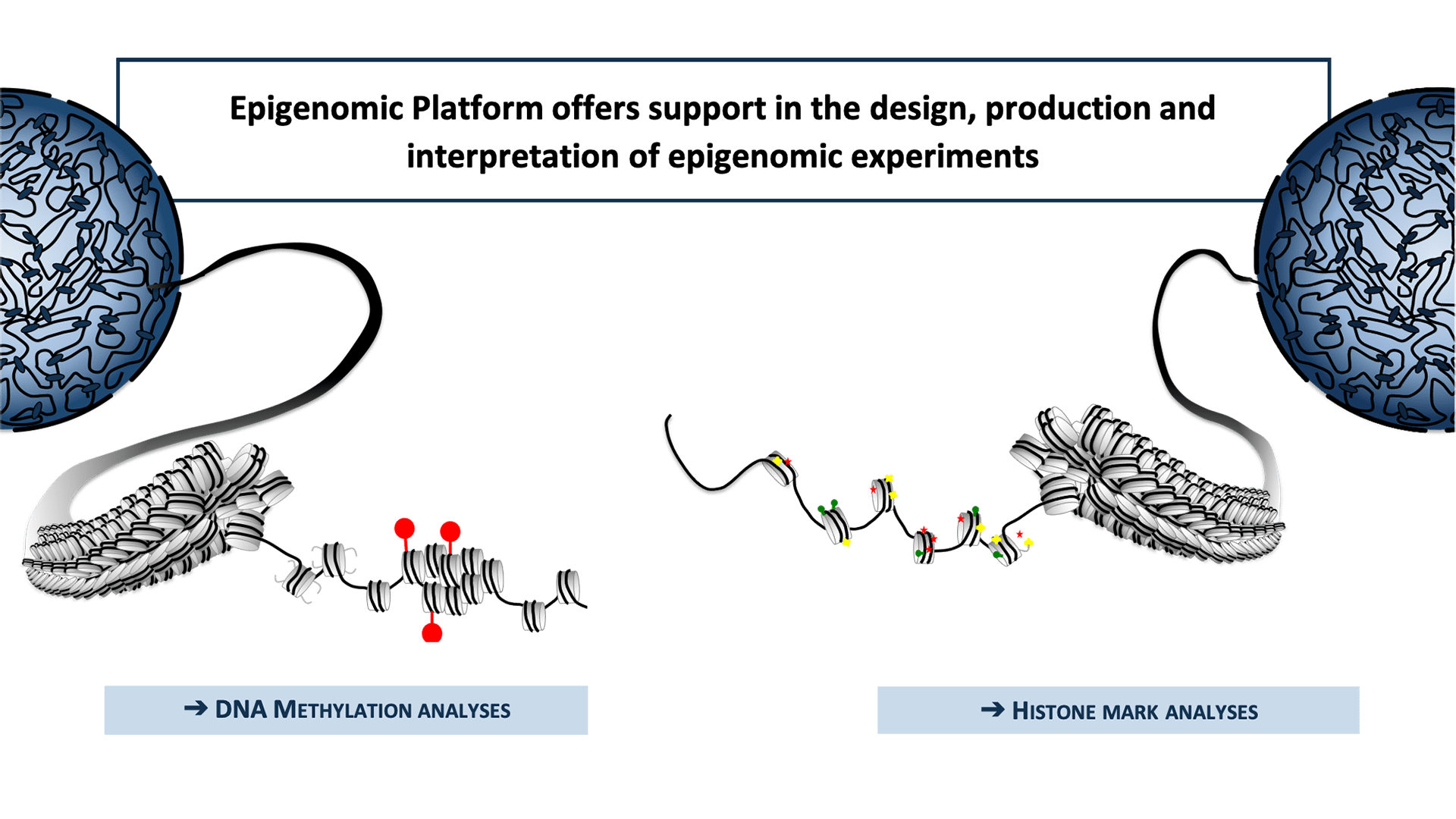
Scheme of platform’s expertise
Epigenomic Platform offers support in the design, production and interpretation of epigenomic experiments.
© Laure Ferry
Expertise
The epigenomic platform offers expertise in analysis of epigenetic marks. It evaluates the feasibility of submitted projects and proposes the most suitable method for their completion.
DNA Methylation
- LUMA (LUminometric Methylation Assay)
- MeDIP ou hMeDIP (Methylated or hydroxyMethylated Immunoprecipitation Assay)
- Bisulfite pyrosequencing
Histones Marks
- Cut & Run
- ChIP
Submit a project
You can contact us for any questions or to discuss your project together.
You must fill in the project request form and send it to the following address: parisepigenetics.pf@gmail.com
Developments
The Functional Epigenomic platform is involved in the development of new techniques for epigenome analysis:
- Cut & Run automatisation
- Cut & Tag
- Enzymatic Methyl-seq ( EM-seq)
- Nanopore
Epigenetic analyses available
Services for DNA methylation and histone marks analyses
Equipment
Equipment available on the Epigenomic platform
Members
Publications
- Planques A, Pierre K, Ferry L, Grunau C, Gazave E, Vervoort M. DNA methylation atlas and machinery in the developing and regenerating annelid Platynereis dumerilii. BMC Biology, Vol 19, Iss 1, Pp 1-26, 2021. DOI: 10.1186/s12915-021-01074-5
- Marchal C, de Dieuleveult M, Saint-Ruf C, Guinot N, Ferry L, Olalla Saad ST, Lazarini M, Defossez PA, Miotto B. Depletion of ZBTB38 potentiates the effects of DNA demethylating agents in cancer cells via CDKN1C mRNA up-regulation. Oncogenesis; Oct 2018, Vol. 7 Issue 10, p1-18, 18p DOI: 10.1038/s41389-018-0092-0
- Velasco G, Grillo G, Touleimat N, Ferry L, Ivkovic I, Ribierre F, Deleuze JF, Chantalat S, Picard C, Francastel C. Comparative methylome analysis of ICF patients identifies heterochromatin loci that require ZBTB24, CDCA7 and HELLS for their methylated state. Human Molecular Genetics, 2018 Jul 15;27(14):2409-2424. DOI: 10.1093/hmg/ddy130.
- Ferry L, Fournier A, Tsusaka T, Adelmant G, Shimazu T, Matano S, Kirsh O, Amouroux R, Dohmae N, Suzuki T, Filion GJ, Deng W, de Dieuleveult M, Fritsch L, Kudithipudi S, Jeltsch A, Leonhardt H, Hajkova P, Marto JA, Arita K, Shinkai Y, Defossez PA. Methylation of DNA Ligase 1 by G9a/GLP Recruits UHRF1 to Replicating DNA and Regulates DNA Methylation. Molecular Cell. August 17, 2017, Vol. 67 Issue 4, p550. DOI: 10.1016/j.molcel.2017.07.012
- Kebir O, Chaumette B, Rivollier F, Miozzo F, Lemieux Perreault LP, Barhdadi A, Provost S, Plaze M, Bourgin J, the ICAAR team, Gaillard R, Mezger V, Dubé M-P , Krebs M-O. Methylomic changes during conversion to psychosis. Molecular Psychiatry (2017) 22, 512-518. DOI: 10.1038/mp.2016.53.
Contact
You can contact us at: parisepigenetics.pf@gmail.com
Functional Epigenomic Platform
35 rue Hélène Brion
Bâtiment Lamarck B, RB77-81
75013 PARIS
+33(0)1 57 27 89 26
Read more

Welcome to Léa
Léa joins the team as a research assistant. After completing a master's degree in virology, she worked in Strasbourg on grapevine viruses, then on characterizing mRNA degradation in plants at the Institute of Plant Molecular Biology (IBMP). In the Polo team, Léa will...

Sophie Polo receives an Impulscience® grant from the Fondation Bettencourt Schueller
Sophie Polo has been awarded an Impulscience® grant to fund a research project on the establishment and maintenance of the inactive X chromosome in response to DNA breaks. This is wonderful news for the lab ! We thank the Fondation Bettencourt Schueller for their...

Welcome to Léa, new engineer in the team!
Léa joins the lab as a research assistant. She holds a Master's degree in Molecular and Cellular Biology from Sorbonne University. She will contribute to investigate DNA methylation maintenance mechanisms in response to UV damage in mammalian cells. Léa Girard À lire...

Well done, Dr Mori!
Margherita successfully defended her PhD on DNA methylation maintenance in response to UV damage. Brava! Margherita and her thesis jury. From left to right: Sophie Polo, Sandra Duharcourt (on screen), Déborah Bourc'his, Margherita Mori, Nataliya Petryk, Jean Molinier,...
