DNA methylation in stem cells
One of our research axes is focused on understanding how DNA methylation patterns are established, maintained, and interpreted during embryonic development. To address these questions we use mouse embryonic stem (ES) cells as our model, and we apply a wide array of tools: cell biology, genomics, proteomics, bioinformatics… This has led us, for example, to decipher important mechanisms of epigenetic memory (Ferry et al, Mol Cell 2017, review in Petryk et al NAR 2020). More recently, we carried out genome-wide CRISPR screens to identify factors linking epigenetics and cellular state (Gupta, Yakhou et al., NSMB 2023).
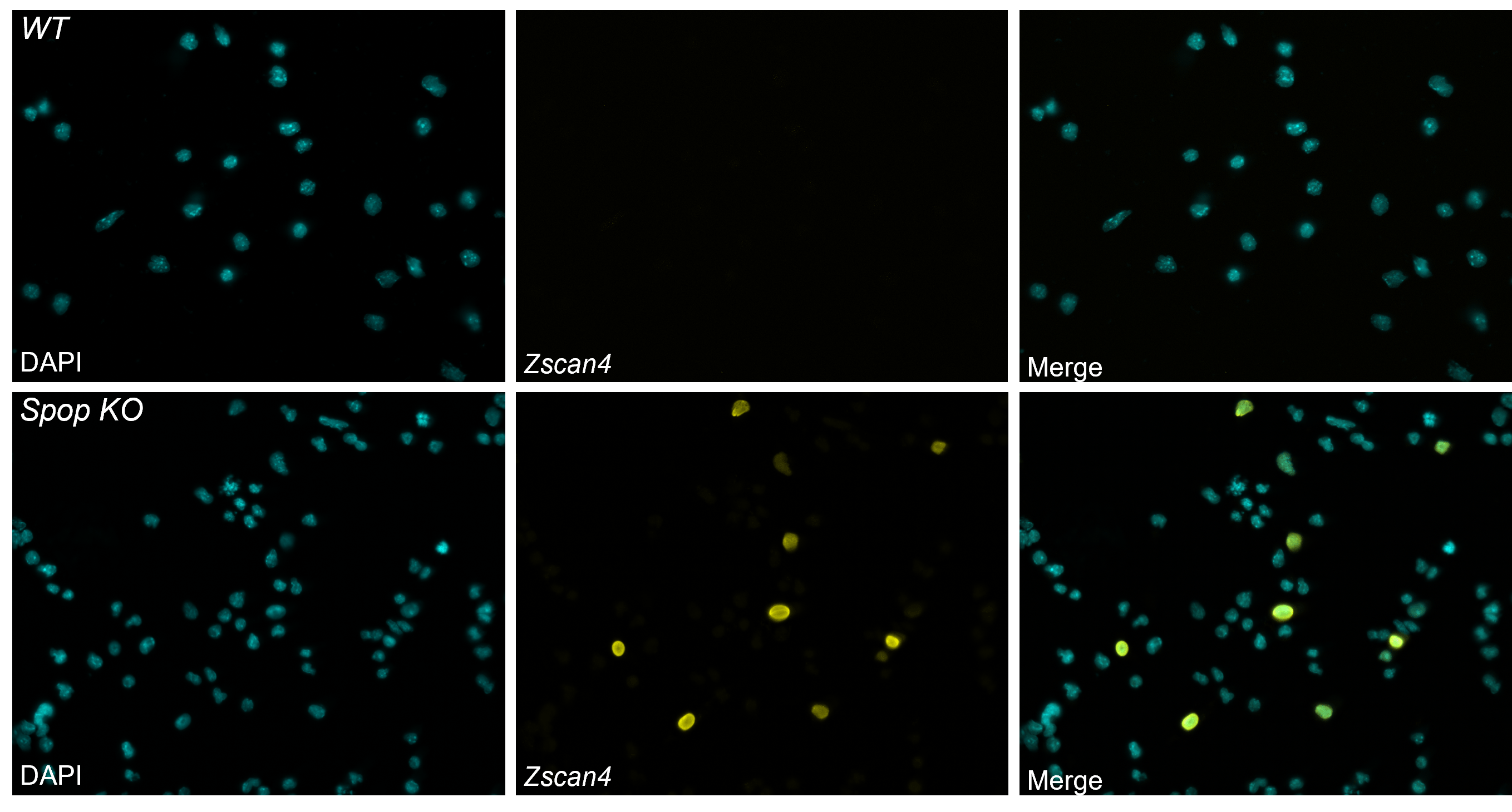
A CRISPR screen identifies new repressors of the totipotent state in mouse embryonic stem cells
In mammals, only the zygote and blastomeres of the early embryo are fully totipotent. This totipotency is reflected in vitro by the expression of markers such as Zscan4. We performed a genome-wide CRISPR KO screen in mouse embryonic stem cells, looking for mutants that reactivate the expression of totipotency markers, like for example the knock-out of Spop.
Read more

Congratulations to Delphine and Julia for their research funding
Congratulations to Delphine Burlet on getting a postdoctoral grant from the Fondation de France and to Julia Roche Dupuy who obtained a fellowship from the HOB doctoral school to finance her PhD thesis. Delphine Burlet, Julia Roche Dupuy À lire aussi

Welcome to Annabelle, new post-doc in the team!
Annabelle joins the lab as a post-doctoral fellow. She completed her PhD at the University of Oxford under the supervision of Prof. Monika Gullerova. Her doctoral work focused on the role of small non-coding Y RNAs and the RNA-binding protein, YBX1, in intracellular...

Welcome to Orlane
Today, we welcome a new member to the team: Orlane. She's a student from the first year of the Brevet de Technicien Supérieur Biotechnologies en Recherche et Production (Two-year technical degree - Biotechnologies in Research and Production).Orlane will be working...

4th year PhD fellowships for Anaëlle
Congratulations to Anaëlle Azogui for obtaining a 4th year PhD fellowships from the "Fondation pour la recherche sur le cancer" (Fondation ARC). Read more
