DNA methylation in stem cells
One of our research axes is focused on understanding how DNA methylation patterns are established, maintained, and interpreted during embryonic development. To address these questions we use mouse embryonic stem (ES) cells as our model, and we apply a wide array of tools: cell biology, genomics, proteomics, bioinformatics… This has led us, for example, to decipher important mechanisms of epigenetic memory (Ferry et al, Mol Cell 2017, review in Petryk et al NAR 2020). More recently, we carried out genome-wide CRISPR screens to identify factors linking epigenetics and cellular state (Gupta, Yakhou et al., NSMB 2023).
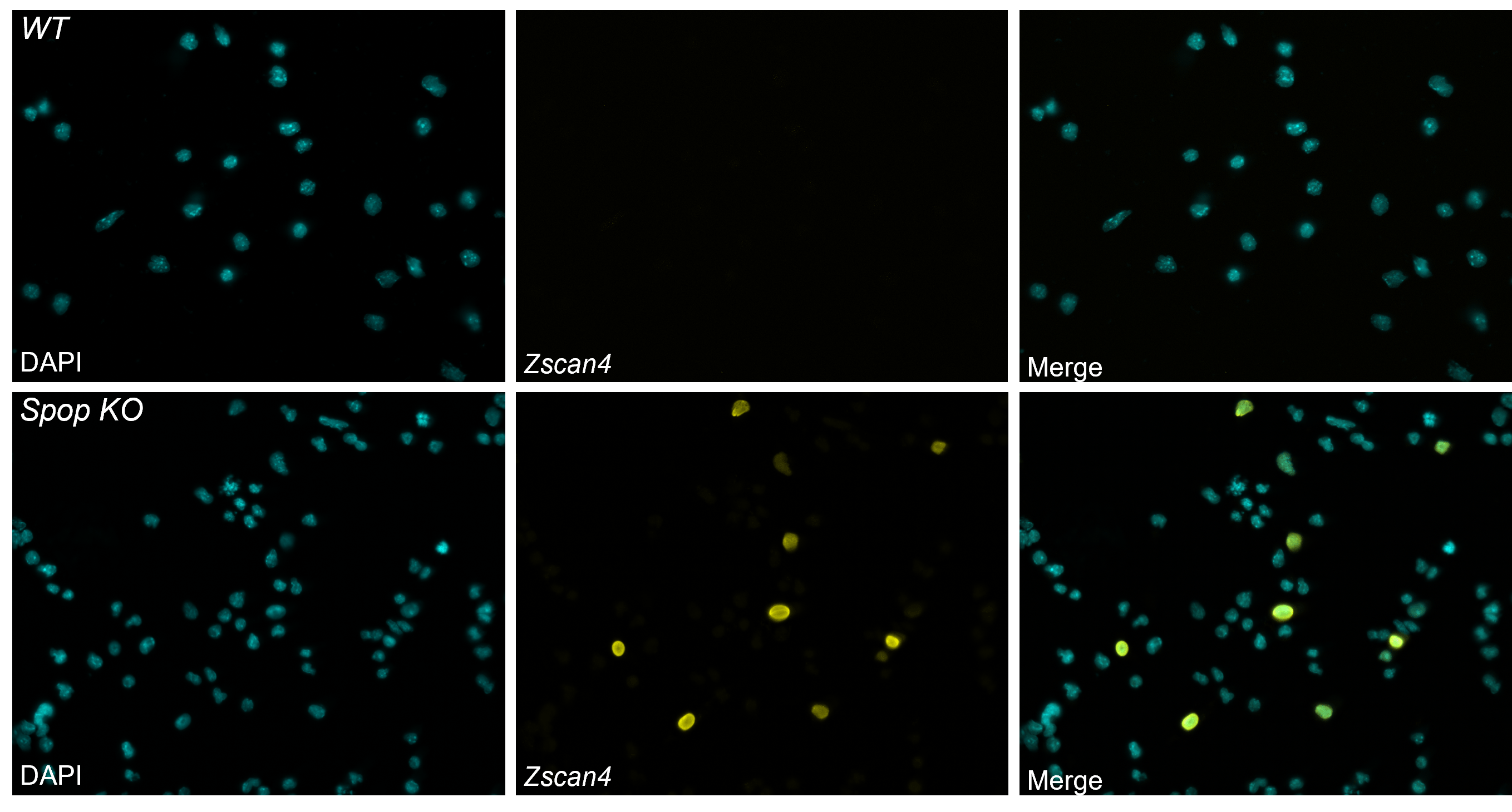
A CRISPR screen identifies new repressors of the totipotent state in mouse embryonic stem cells
In mammals, only the zygote and blastomeres of the early embryo are fully totipotent. This totipotency is reflected in vitro by the expression of markers such as Zscan4. We performed a genome-wide CRISPR KO screen in mouse embryonic stem cells, looking for mutants that reactivate the expression of totipotency markers, like for example the knock-out of Spop.
Read more

Welcome to Léa
Léa joins the team as a research assistant. After completing a master's degree in virology, she worked in Strasbourg on grapevine viruses, then on characterizing mRNA degradation in plants at the Institute of Plant Molecular Biology (IBMP). In the Polo team, Léa will...

Sophie Polo receives an Impulscience® grant from the Fondation Bettencourt Schueller
Sophie Polo has been awarded an Impulscience® grant to fund a research project on the establishment and maintenance of the inactive X chromosome in response to DNA breaks. This is wonderful news for the lab ! We thank the Fondation Bettencourt Schueller for their...

Welcome to Léa, new engineer in the team!
Léa joins the lab as a research assistant. She holds a Master's degree in Molecular and Cellular Biology from Sorbonne University. She will contribute to investigate DNA methylation maintenance mechanisms in response to UV damage in mammalian cells. Léa Girard À lire...

Well done, Dr Mori!
Margherita successfully defended her PhD on DNA methylation maintenance in response to UV damage. Brava! Margherita and her thesis jury. From left to right: Sophie Polo, Sandra Duharcourt (on screen), Déborah Bourc'his, Margherita Mori, Nataliya Petryk, Jean Molinier,...
