Centromeric repeats transcription
A paradigm to link transcription of DNA repeats to global molecular and cellular effects
Cells in several phases of the cell cycle
© EDC
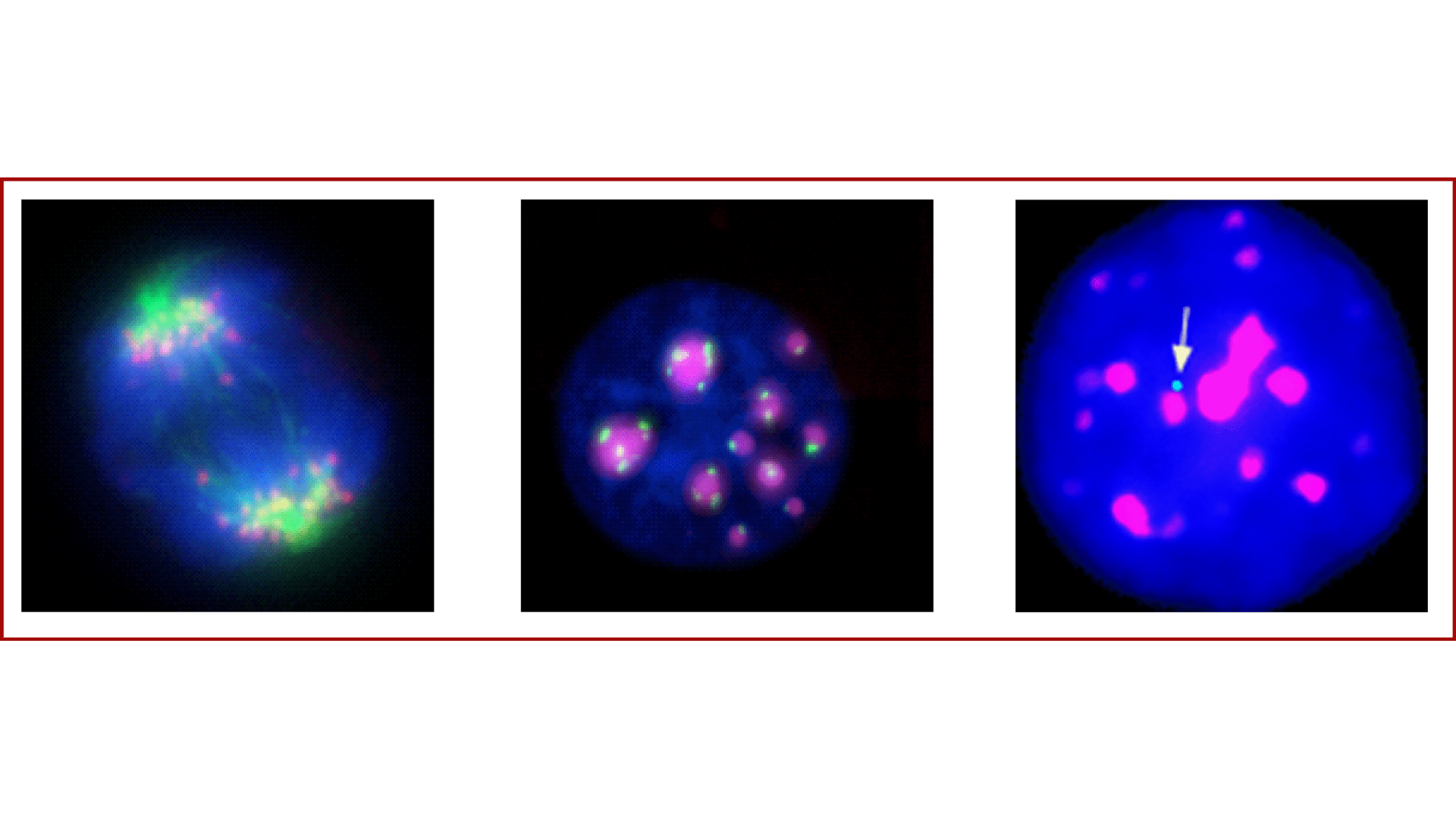
Tandem repeats that underlie centromeric regions have a structural role at the chromosomal level, providing the assembly platforms for the kinetochore and attachment of the mitotic spindle, but also in the functional organization of the nucleus and long-range control of genome expression.
We characterized transcripts that originate from murine centromeric repeats and showed that they are essential for centromere identity and function, whereas their unscheduled accumulation is causally linked to perturbed nuclear organization and cellular phenotypes. However, we showed that the outcome greatly depends on cellular and genetic contexts. In primary cells, increased transcription of centromeric repeats functions as a sensor of stress promoting cell cycle arrest and safeguard mechanisms; in contrast, in contexts of loss of the p53 checkpoints it leads to chromosomal instability.
We explore the causal link between aberrant transcription of repetitive sequences and perturbed molecular and cellular programs, ex vivo in various cellular and genotype context and in mouse models. We also question the mechanisms that lead to their deregulated transcription, with special interest on DNA methylation that is tightly linked with maintenance of integrity of these sequences and hence, with maintenance of genome stability. Ultimately, we aim at deciphering the cellular functions and regulatory factors deregulated by their unscheduled accumulation.
Related publications:
- Hédouin S, Grillo G, Ivkovic I, Velasco G, Francastel C. CENP-A chromatin disassembly in stressed and senescent murine cells. Sci Rep. 2017 Feb 10;7:42520. doi: 10.1038/srep42520. PMID: 28186195
- Guillemin C, Francastel C. [Heterochromatin compartments and gene silencing: human hematopoietic differentiation as a model study]. Biol Aujourdhui. 2010;204(3):221-33. Review. French. PMID: 20950566
- Guillemin C, Maleszewska M, Guais A, Maës J, Rouyez MC, Yacia A, Fichelson S, Goodhardt M, Francastel C. Chromatin modifications in hematopoietic multipotent and committed progenitors are independent of gene subnuclear positioning relative to repressive compartments. Stem Cells. 2009 Jan;27(1):108-15. PMID: 18974210
- Ferri F, Bouzinba-Segard H, Velasco G, Hubé F, Francastel C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009 Aug;37(15):5071-80. PMID: 19542185
Read more

Welcome to Léa
Léa joins the team as a research assistant. After completing a master's degree in virology, she worked in Strasbourg on grapevine viruses, then on characterizing mRNA degradation in plants at the Institute of Plant Molecular Biology (IBMP). In the Polo team, Léa will...

Sophie Polo receives an Impulscience® grant from the Fondation Bettencourt Schueller
Sophie Polo has been awarded an Impulscience® grant to fund a research project on the establishment and maintenance of the inactive X chromosome in response to DNA breaks. This is wonderful news for the lab ! We thank the Fondation Bettencourt Schueller for their...

Welcome to Léa, new engineer in the team!
Léa joins the lab as a research assistant. She holds a Master's degree in Molecular and Cellular Biology from Sorbonne University. She will contribute to investigate DNA methylation maintenance mechanisms in response to UV damage in mammalian cells. Léa Girard À lire...

Well done, Dr Mori!
Margherita successfully defended her PhD on DNA methylation maintenance in response to UV damage. Brava! Margherita and her thesis jury. From left to right: Sophie Polo, Sandra Duharcourt (on screen), Déborah Bourc'his, Margherita Mori, Nataliya Petryk, Jean Molinier,...
