Alternative splicing of introns and the diversification of transcriptional output
A paradigm to test the functionality of non-protein-coding genomic regions
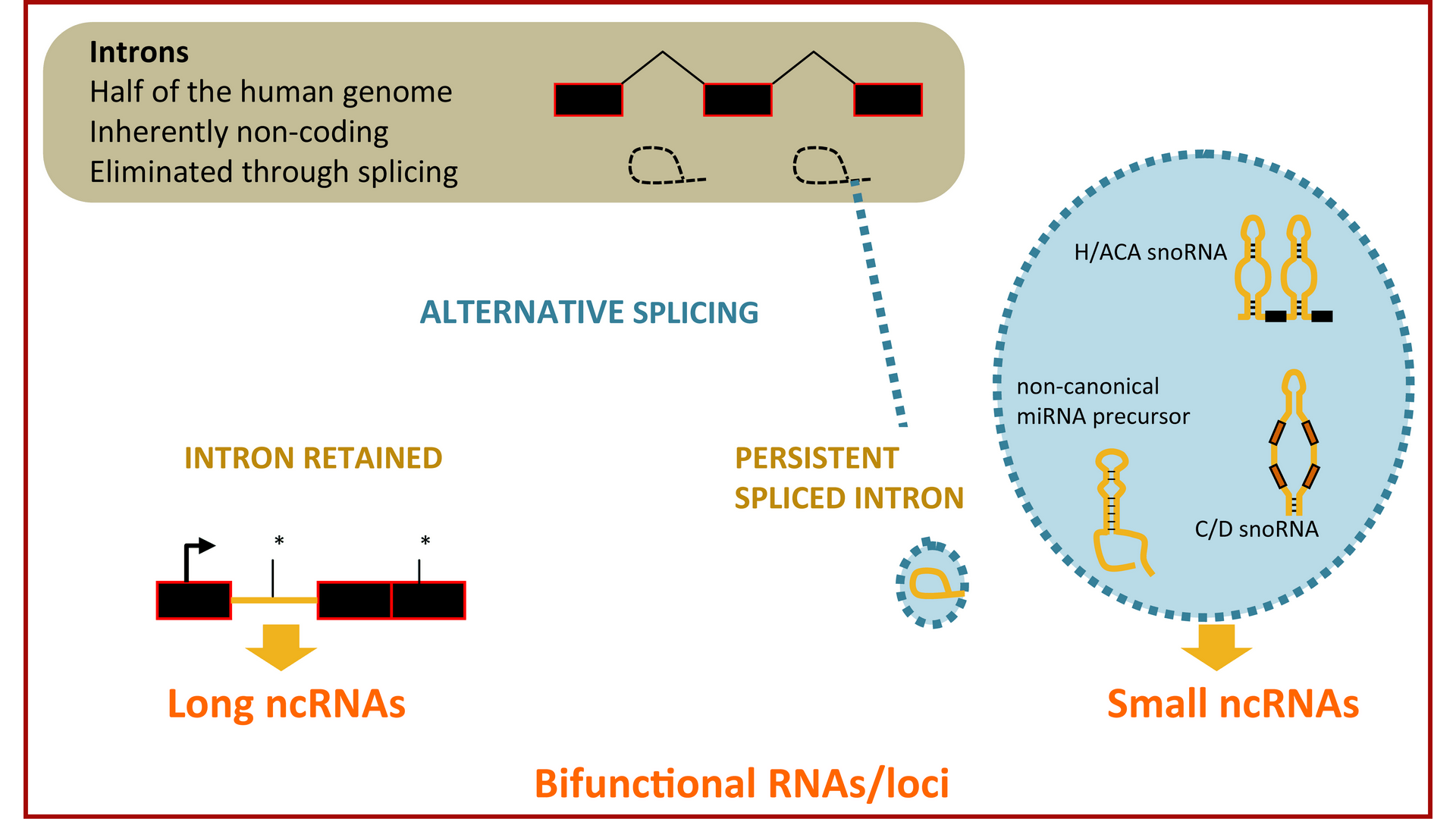
Bifunctional RNA
© EDC
Introns represent almost half of the human genome, although their vast majority is eliminated from eukaryotic transcripts through RNA splicing. Yet, important information is embedded within introns, the most remarkable being the release of different classes of small regulatory ncRNAs directly from splicing. In addition, we showed that their excision or retention, depending on cellular context, contributes to the diversification of the information carried by genes by producing functional RNAs instead of a protein-coding mRNA.
Hence, alternative splicing is a versatile developmental switch that provides plasticity to eukaryotic genomes transcriptional output, increasing not only proteome but also transcriptome diversity.
We explore alternative splicing of introns as a mechanism to fine-tune the production of long and short intron-derived regulatory ncRNAs (that we termed SID) during normal and pathological muscle differentiation where splicing is impaired like in Myotonic Dystrophy type 1, through bioinformatics predictions, high-throughput RNA sequencing and functional validation in model systems.
Related publications:
- Bogard B*, Tellier G*, Francastel C, Hubé F. A Tool to Design Bridging Oligos Used to Detect Pseudouridylation Sites on RNA after CMC Treatment. Non-coding RNA, 2022, 8(5):63. PMID: 36287115
- Bogard B, Francastel C, Hubé F. Multiple information carried by RNAs: total eclipse or a light at the end of the tunnel? RNA Biol, 2020, 17(12):1707-1720. PMID: 32559119
- Hubé F, Francastel C. Coding and non-coding RNAs, the frontier has never been so blurred. Frontiers in Genetics, 2018, 9:140. PMID: 29720998
- Bogard B, Francastel C, Hubé F. A new method for the identification of thousands of circular RNAs. Non-coding RNA Investig, 2018, 2:5. Site: Fulltext
- Hubé F, Ulveling D, Sureau A, Forveille S, Francastel C. Short intron-derived ncRNAs. Nucleic Acids Res. 2017 Jan 3;45(8):4768-4781. PMID: 28053119
- Hubé F, Francastel C. “Pocket-sized RNA-Seq”: a method to capture new mature microRNA produced from a genomic region of interest. Non-coding RNA. 2015 June;1(2):127-138. Paper here
- Hubé F, Francastel C. Mammalian Introns: When the Junk Generates Molecular Diversity. Int J of Mol Sci. 2015 Feb 20;16(3):4429-4452. PMID: 25710723
- Ulveling D, Dinger M, Francastel C, Hubé F. Identification of a dinucleotide signature that discriminates coding from non-coding long RNAs. Front Genet. 2014 Sep 9;5:316. PMID: 25250049
- Ulveling D, Francastel C, Hubé F. When one is better than two: RNA with dual functions. Biochimie. 2011 Apr;93(4):633-44. Review. PMID: 21111023
- Ulveling D, Francastel C, Hubé F. Identification of potentially new bifunctional RNA based on genome-wide data-mining of alternative splicing events. Biochimie. 2011 Nov;93(11):2024-7. PMID: 21729736
- Hubé F, Velasco G, Rollin J, Furling D, Francastel C. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 2011 Jan;39(2):513-25. PMID: 20855289
Read more

Congratulations to Delphine and Julia for their research funding
Congratulations to Delphine Burlet on getting a postdoctoral grant from the Fondation de France and to Julia Roche Dupuy who obtained a fellowship from the HOB doctoral school to finance her PhD thesis. Delphine Burlet, Julia Roche Dupuy À lire aussi

Welcome to Annabelle, new post-doc in the team!
Annabelle joins the lab as a post-doctoral fellow. She completed her PhD at the University of Oxford under the supervision of Prof. Monika Gullerova. Her doctoral work focused on the role of small non-coding Y RNAs and the RNA-binding protein, YBX1, in intracellular...

Welcome to Orlane
Today, we welcome a new member to the team: Orlane. She's a student from the first year of the Brevet de Technicien Supérieur Biotechnologies en Recherche et Production (Two-year technical degree - Biotechnologies in Research and Production).Orlane will be working...

4th year PhD fellowships for Anaëlle
Congratulations to Anaëlle Azogui for obtaining a 4th year PhD fellowships from the "Fondation pour la recherche sur le cancer" (Fondation ARC). Read more
