EpHISTain : Histology and cytology
The EpHISTain facility supports the teams of the “Epigenetics and Cell Fate” Unit for the tissus and cells phenotyping

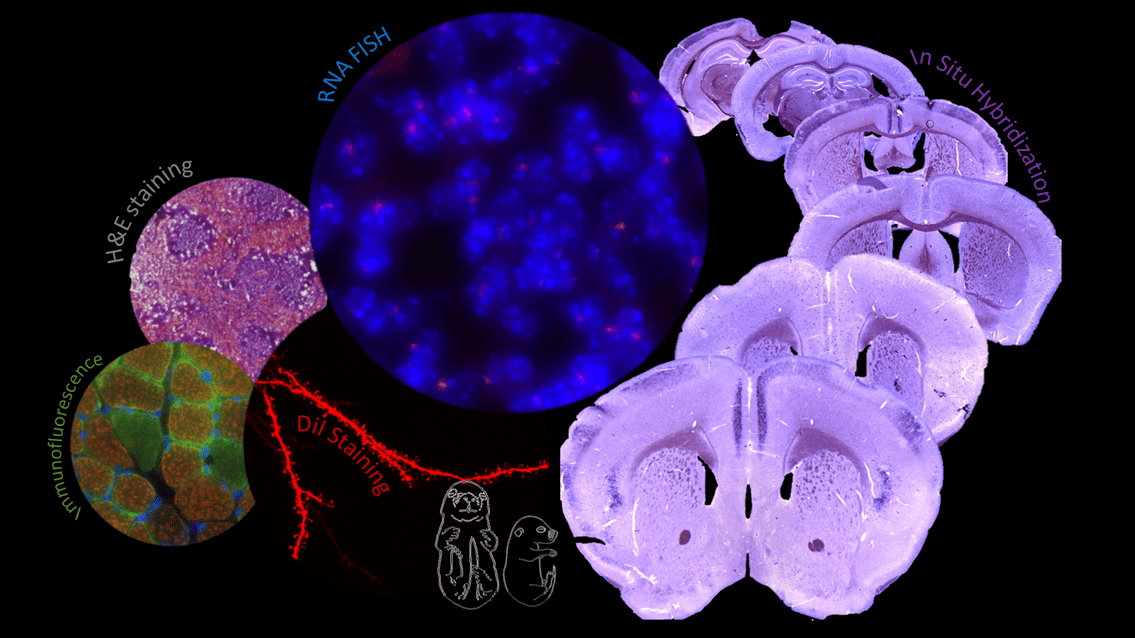
Illustration of experiments carried out by the EpHISTain platform showing from left to right:
Immunofluorescence on mouse muscle section, DiI neuron labelling on adult mouse brain section, H&E staining on mouse spleen section, RNA FISH on mouse brain section, in situ hybridization on mouse brain section.
© Myriame MOHAMED
Presentation
EpHISTain facility is dedicated to the technical support of the teams of the “Epigenetics and Cell Fate” Unit in the fields of histology and cytology in order to support the research work of the teams in the elucidation of the role, the mechanisms of implementation and maintenance of epigenetic modifications that determine plasticity and cell fate.
Expertises
This service sets up various histological techniques, histochemistry, immunohistochemistry, immunofluorescence, in situ hybridization (ISH), fluorescence in situ hybridization (FISH) and cytology, for biological tissue sections production and staining (animal or organoid), as well as cells labelling.
Services
- Technical support or sample handling (fresh, fixed, frozen) for the biological tissue sections production. The chosen technique depends on the technical and scientific teams’ questions, as well as the treatments necessary to answer them:
- Tissue dissection, preparation and fixation (including embryonic)
- Tissue embedding (OCT and agarose)
- Tissue sectioning (vibratome or cryostat), on slides or in floating sections
- Biological tissue sections treatment. The treatment choice depends on the type of tissue, the technical issues and the biological question to be answered by the experiment:
- Explore tissues morphology or the structures organization of interest on sections in order to highlight possible modifications or structural defects
-
- Histochemical staining on biological tissue sections (e.g. cresyl violet)
- In Situ Hybridization (ISH) on cryostat sections, floating or on slides, for the labelling of specific structures (e.g.: cortical layers markers)
- Characterize the expression profile of genes of interest (expression or not and cellular localization) by targeting RNAs or proteins
-
- In Situ Hybridization (ISH) on cryostat sections, floating or on slides
- Immunohistochemistry and Immunofluorescence
- FISH (DNA & RNA)
- Imaging quality control of results (on the EPI2, EDC platform)
- Support
- Optimize and standardize the labeling protocols on tissues and cells for homogeneity and reproducibility which allow the teams’ data to be crossed.
- Adapt and develop staining protocols on tissues or cells according to the specific questions of the teams, by providing technical expertise
- Train team members in sectionning and labelling techniques as needed, sharing protocols and know-how.
Members
Myriame MOHAMED
Head, TCS CNRS
Steering Committee
- Véronique Dubreuil, Assistant Professor (Université Paris Cité)
- Céline Morey, Senior Researcher (INSERM)
Contacts
For all information requests, please contact the EpHISTain platform:
Myriame MOHAMED, TCS CNRS
Epigenetics and Cell Fate Center
CNRS UMR7216 – Université Paris Cité
Lamarck B – 4th floor – room 444
35, rue Hélène Brion
75205 Paris Cedex 13
Email : myriame.mohamed@u-paris.fr
Tel : 33 (0)1 57 27 89 25
Fax : 33 (0)1 57 27 89 11
À lire aussi

Welcome to Léa
Léa joins the team as a research assistant. After completing a master's degree in virology, she worked in Strasbourg on grapevine viruses, then on characterizing mRNA degradation in plants at the Institute of Plant Molecular Biology (IBMP). In the Polo team, Léa will...

Sophie Polo receives an Impulscience® grant from the Fondation Bettencourt Schueller
Sophie Polo has been awarded an Impulscience® grant to fund a research project on the establishment and maintenance of the inactive X chromosome in response to DNA breaks. This is wonderful news for the lab ! We thank the Fondation Bettencourt Schueller for their...

Welcome to Léa, new engineer in the team!
Léa joins the lab as a research assistant. She holds a Master's degree in Molecular and Cellular Biology from Sorbonne University. She will contribute to investigate DNA methylation maintenance mechanisms in response to UV damage in mammalian cells. Léa Girard À lire...

Well done, Dr Mori!
Margherita successfully defended her PhD on DNA methylation maintenance in response to UV damage. Brava! Margherita and her thesis jury. From left to right: Sophie Polo, Sandra Duharcourt (on screen), Déborah Bourc'his, Margherita Mori, Nataliya Petryk, Jean Molinier,...
