Epigenetic analyses available
Depending on the question of interest, the Epigenomics platform offers different techniques for the analysis of DNA methylation and histone marks.
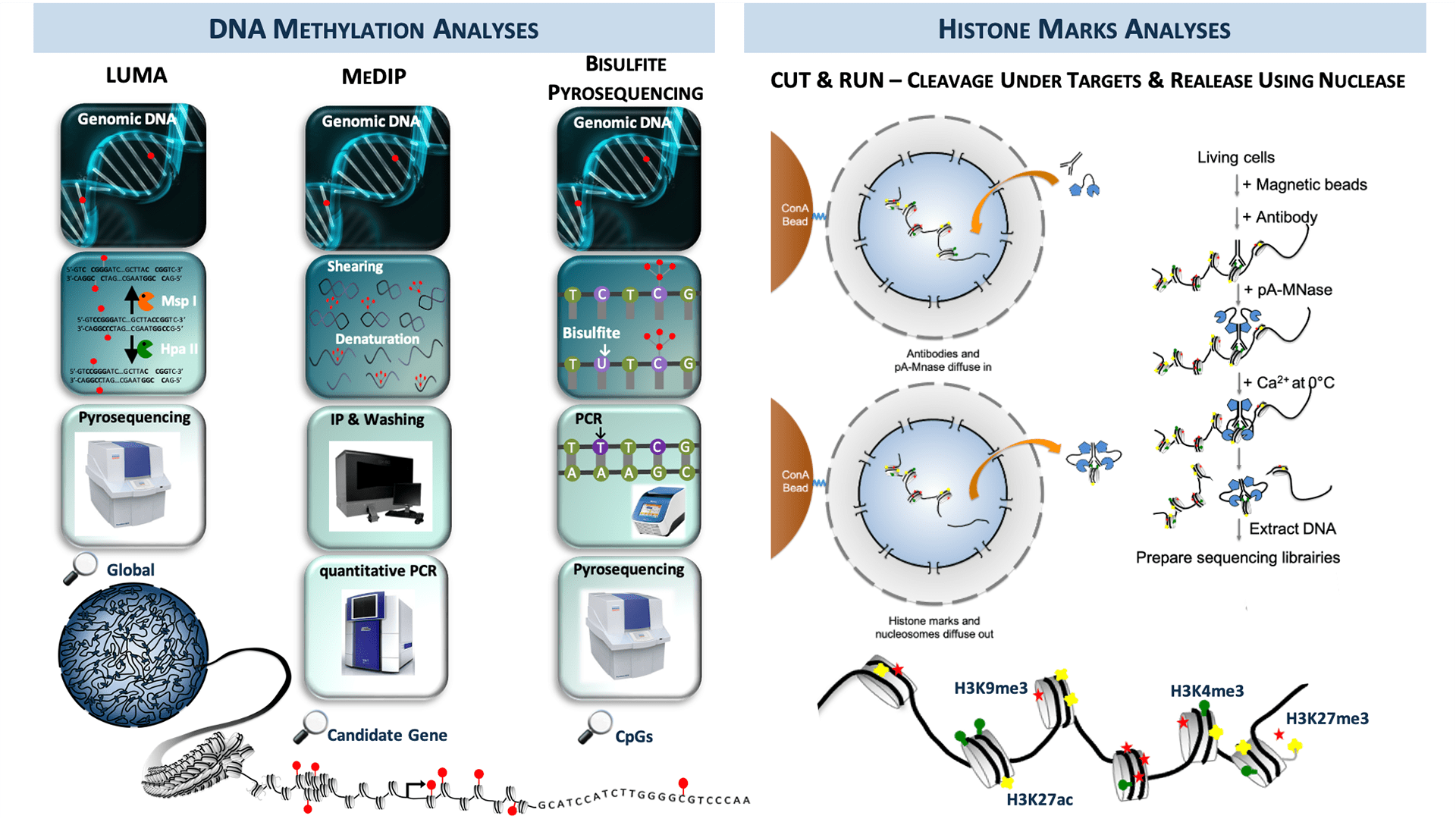
Scheme of the techniques proposed on the Epigenomic platform.
The Epigenomic platform offers techniques for the analysis of DNA methylation and histone marks.
© Laure Ferry
DNA Methylation analyses
LUMA (LUminometric Methylation Assay)
- Method for global DNA methylation analysis.
- Quantification of the number of restriction enzyme cuts, based on methylation sensitivity.
- Rapid & quantitative determination of methylation level.
- No information about the location or repartition of DNA methylation.
MeDIP (Methylated DNA ImmunoPrécipitation)
-
Enrichment of fragmented genomic DNA using an antibody specific for 5mC
-
Low CpG dinucleotide density may not be accurately analyzed by MeDIP.
-
5mC antibodies are capable of discrimination between 5-hmC and 5-mC.
Bisulfite Pyrosequencing
-
Analysis & quantification of the methylation degree of several CpG positions in close proximity.
-
Bisulfite-based methods cannot differentiate 5-mC and 5-hmC.
Histones Marks analyses
CUT & RUN (Cleavage Under Targets & Release Using Nuclease)
-
Powerful and versatile technique used for probing protein-DNA interactions within the natural chromatin context of the cell
-
CUT&RUN provides a rapid, robust with low cell number assay
-
Free from formaldehyde cross-linking, chromatin fragmentation, and immunoprecipitation, making it a more efficient method for identifying target genes.
Read more

Welcome to Léa
Léa joins the team as a research assistant. After completing a master's degree in virology, she worked in Strasbourg on grapevine viruses, then on characterizing mRNA degradation in plants at the Institute of Plant Molecular Biology (IBMP). In the Polo team, Léa will...

Sophie Polo receives an Impulscience® grant from the Fondation Bettencourt Schueller
Sophie Polo has been awarded an Impulscience® grant to fund a research project on the establishment and maintenance of the inactive X chromosome in response to DNA breaks. This is wonderful news for the lab ! We thank the Fondation Bettencourt Schueller for their...

Welcome to Léa, new engineer in the team!
Léa joins the lab as a research assistant. She holds a Master's degree in Molecular and Cellular Biology from Sorbonne University. She will contribute to investigate DNA methylation maintenance mechanisms in response to UV damage in mammalian cells. Léa Girard À lire...

Well done, Dr Mori!
Margherita successfully defended her PhD on DNA methylation maintenance in response to UV damage. Brava! Margherita and her thesis jury. From left to right: Sophie Polo, Sandra Duharcourt (on screen), Déborah Bourc'his, Margherita Mori, Nataliya Petryk, Jean Molinier,...
